Kbay Bleaching 2019 DNA RNA Extractions
DNA RNA Duet Extractions for Kbay Bleaching 2019 Project
Github project link.
Raw molecular lab work data spreadsheet link.
Coral metadata spreadsheet link.
Google drive for extraction photos link.
Protocol
Based on Putnam Lab DNA RNA Duet Extraction Protocol and Zymo Research Kit.
After clipping fragments and placing them into the sample tube. The goal is to have all the samples start with this volume of coral.

After 2 minute vortex step, this is what the sample tube will look like. The goal is to have almost all of the tissue off of the coral skeleton. Montipora spp. can be difficult to do this without resulting in lower quality DNA/RNA because they are so mucus-y.
September, October, and December 2019 Extractions
2021-01-31 Extractions (1-6)
Qubit Standards:
DNA BR St1: 180.18 St2: 19,569.74
RNA HS St1: 46.32 St2: 728.78
| ColonyID | Collection-Date | Bleach- | Extraction-ID | Extraction-Date | DNA reading 1 (ng/uL) | DNA reading 2 (ng/uL) | DNA reading Avg (ng/uL) | RNA reading 1 (ng/uL) | RNA reading 2 (ng/uL) | RNA reading Avg (ng/uL) | RIN | Gel Pass? | Final Extraction for this coral? | Notes |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M-204 | 2019-10-02 | Non-Bleach | 1 | 20210131 | 5.84 | 5.78 | 5.81 | 43.2 | 43 | 43.1 | 3.4 | No | NA | Degraded |
| M-201 | 2019-12-04 | Bleach | 2 | 20210131 | 20.4 | 20.2 | 20.3 | 22 | 22 | 22 | 8.5 | Yes | Yes | NA |
| M-212 | 2019-10-02 | Non-Bleach | 3 | 20210131 | 7.06 | 7.04 | 7.05 | 66.4 | 66.4 | 66.4 | 2.6 | No | NA | Degraded |
| M-11 | 2019-12-04 | Bleach | 4 | 20210131 | 14.7 | 14.6 | 14.65 | 21.4 | 21.4 | 21.4 | 8.6 | Yes | Yes | NA |
| M-203 | 2019-10-02 | Bleach | 5 | 20210131 | 57.8 | 57.6 | 57.7 | 36.6 | 36.6 | 36.6 | 6 | No | NA | Degraded |
| M-202 | 2019-12-04 | Non-Bleach | 6 | 20210131 | 24.6 | 24.4 | 24.5 | 21.4 | 21.2 | 21.3 | 8.2 | Yes | Yes | NA |
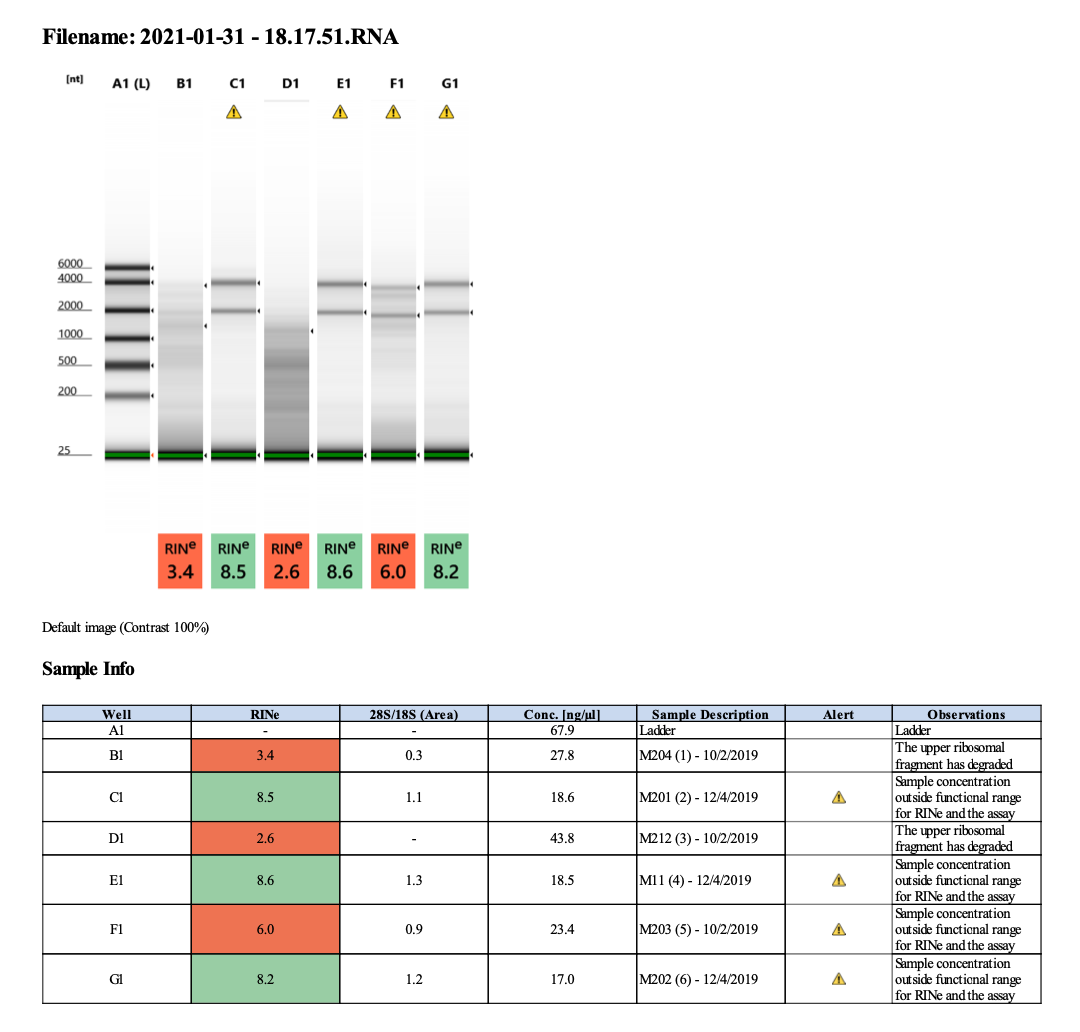
Full tapestation report 2021-01-31.
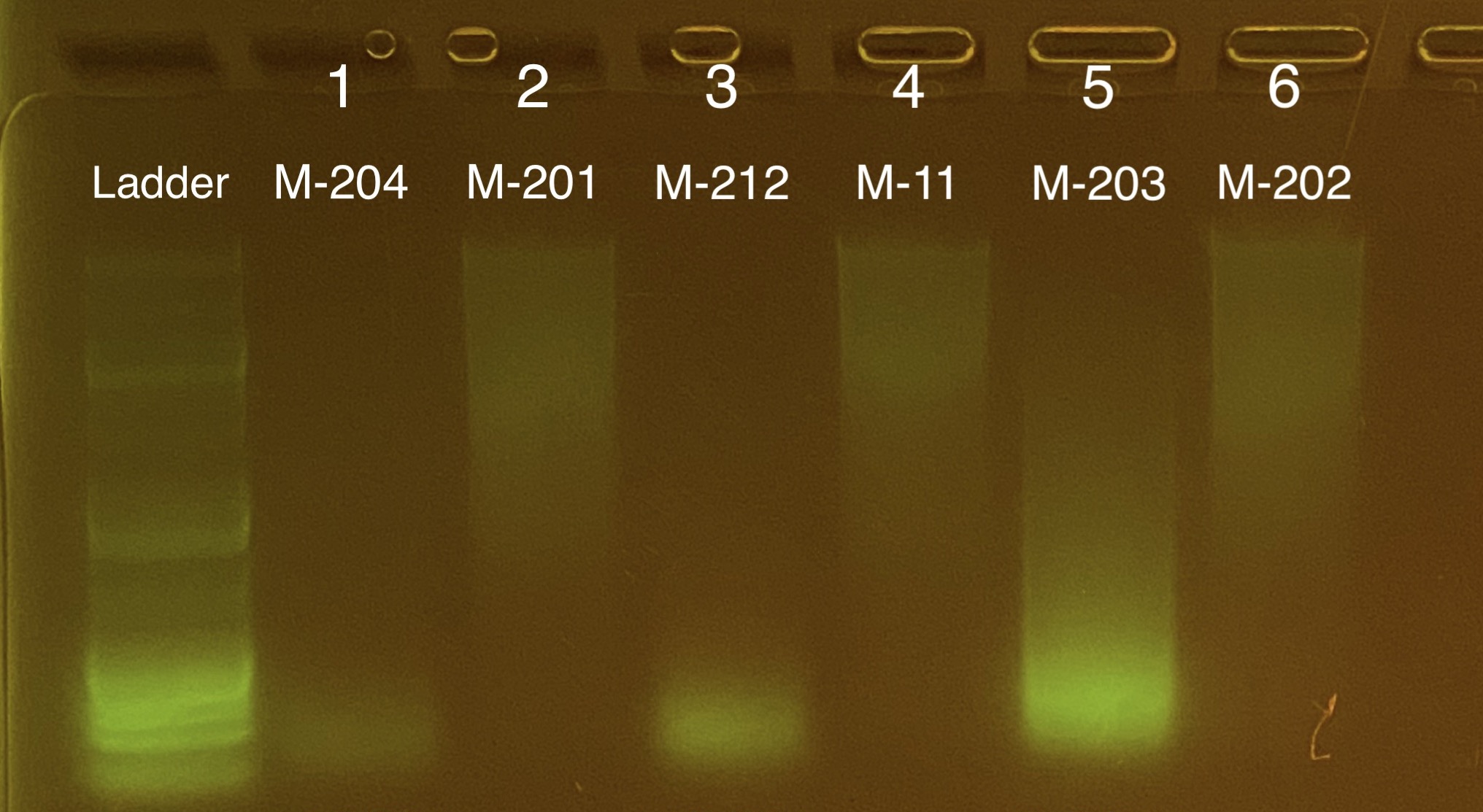
All Oct 2 2019 samples are completely degraded. These samples likely thawed in transit at some point. Dec 4 2019 look great.
2021-02-10 Extractions (7-12)
Since Oct 2 2019 was degraded, we wanted to try to extraction from 2 corals from Sept 2019, Oct 16 2019, and Oct 30 2019. All four timepoints from the Fall of 2019 are degraded: 2019-09-16, 2019-10-02, 2019-10-16, and 2019-10-30.
Qubit Standards: DNA BR St1: 193.97 St2: 22,049.17 RNA HS St1: 48.07 St2: 810.29
| ColonyID | Collection-Date | Bleach- | Extraction-ID | Extraction-Date | DNA reading 1 (ng/uL) | DNA reading 2 (ng/uL) | DNA reading Avg (ng/uL) | RNA reading 1 (ng/uL) | RNA reading 2 (ng/uL) | RNA reading Avg (ng/uL) | RIN | Gel Pass? | Final Extraction for this coral? | Notes |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M-3 | 2019-10-30 | Bleach | 7 | 20210210 | 17.3 | 17.2 | 17.25 | 31.2 | 31 | 31.1 | 6.3 | No | NA | Degraded |
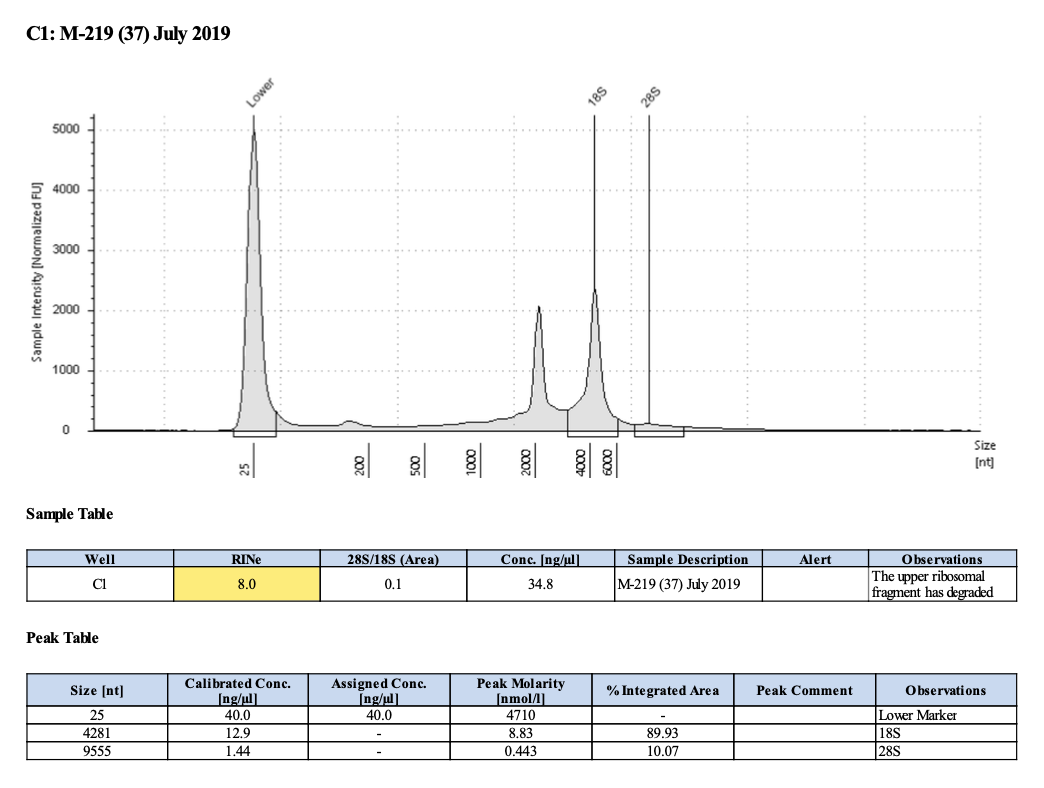
| M-219 | 2019-10-30 | Bleach | 8 | 20210210 | 20.6 | 20.6 | 20.6 | 22.2 | 22.2 | 22.2 | 7.3 | No | NA | Degraded |
| M-218 | 2019-10-16 | Non-Bleach | 9 | 20210210 | 8.08 | 8.02 | 8.05 | 45.6 | 45.6 | 45.6 | 2.9 | No | NA | Degraded |
| M-202 | 2019-10-16 | Non-Bleach | 10 | 20210210 | 6.78 | 6.76 | 6.77 | 15.3 | 15.3 | 15.3 | 3.2 | No | NA | Degraded |
| M-218 | 2019-09-16 | Non-Bleach | 11 | 20210210 | 4.48 | 4.44 | 4.46 | 54 | 53.8 | 53.9 | 3.2 | No | NA | Degraded |
| M-201 | 2019-09-16 | Bleach | 12 | 20210210 | 21 | 21 | 21 | 37.8 | 37.8 | 37.8 | 5.3 | No | NA | Degraded |
Gel order: Ladder, 7, 8, 9, 10, 11, 12.
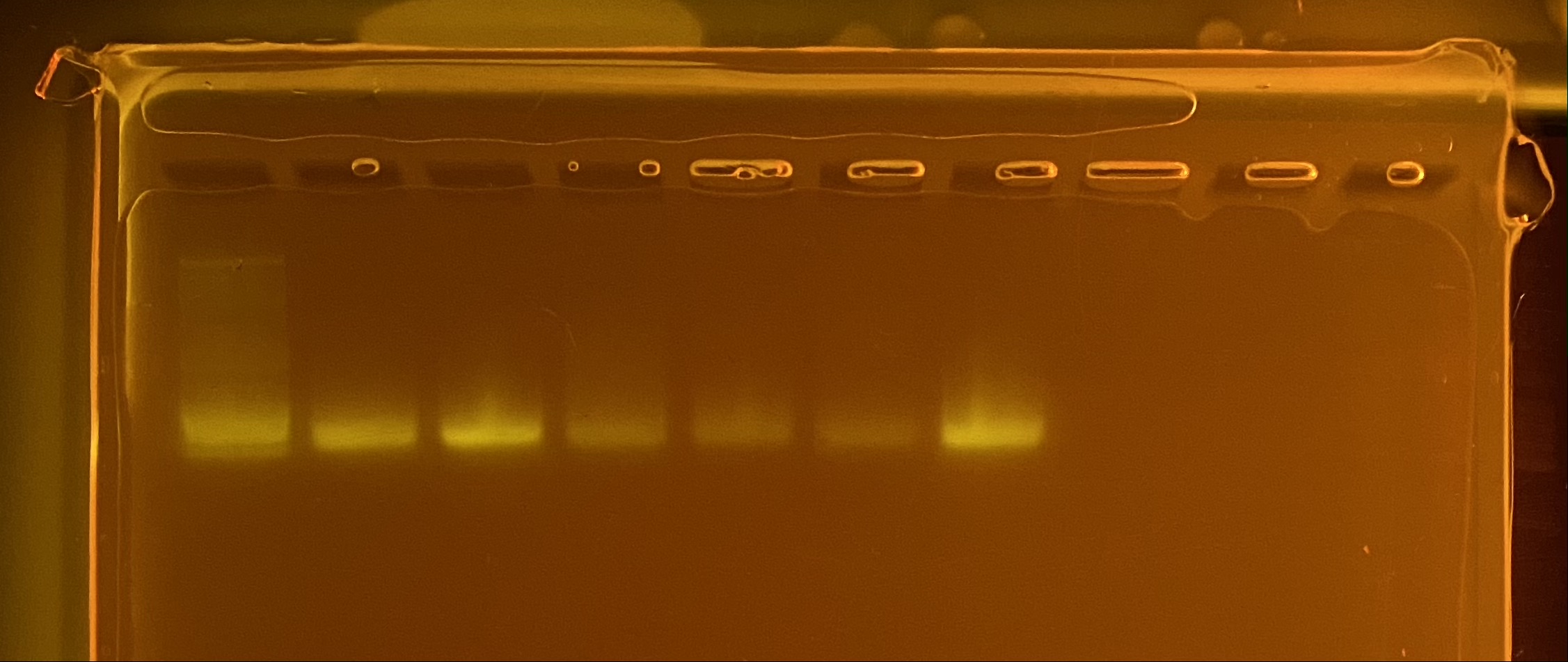
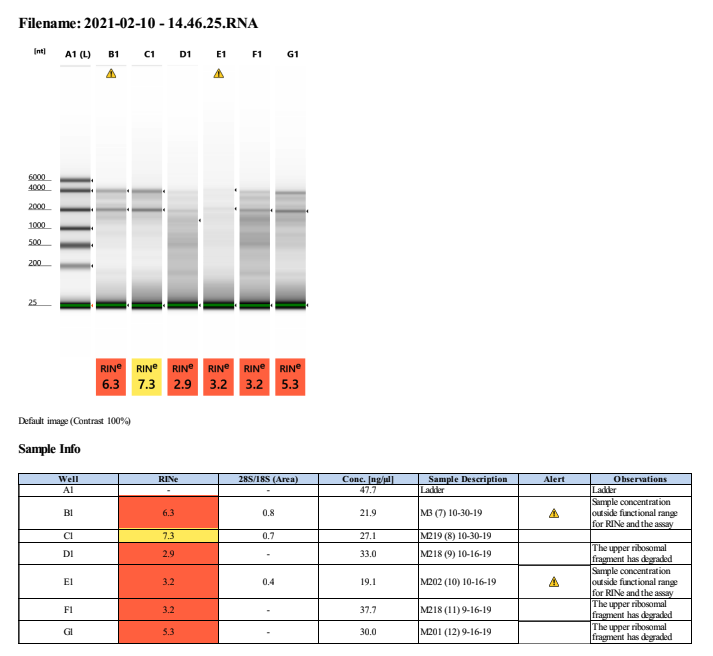
Full tapestation report found here.
20210211 Nanodrop 12-4-2019 samples
Nandropped three M. capitata samples in the GSC.
The ratio of absorbance at 260 nm and 280 nm is used to assess the purity of DNA and RNA. A ratio of ~1.8 is generally accepted as “pure” for DNA; a ratio of ~2.0 is generally accepted as “pure” for RNA.
This ratio is used as a secondary measure of nucleic acid purity. The 260/230 values for “pure” nucleic acid are often higher than the respective 260/280 values. Expected 260/230 values are commonly in the range of 2.0-2.2. If the ratio is appreciably lower than expected, it may indicate the presence of contaminants which absorb at 230 nm. EDTA, carbohydrates and phenol all have absorbance near 230 nm.
| Extraction-ID | Extraction-Date | ColonyID | Collection-Date | Bleach-Status | 260/280 | 260/230 | ng_uL |
|---|---|---|---|---|---|---|---|
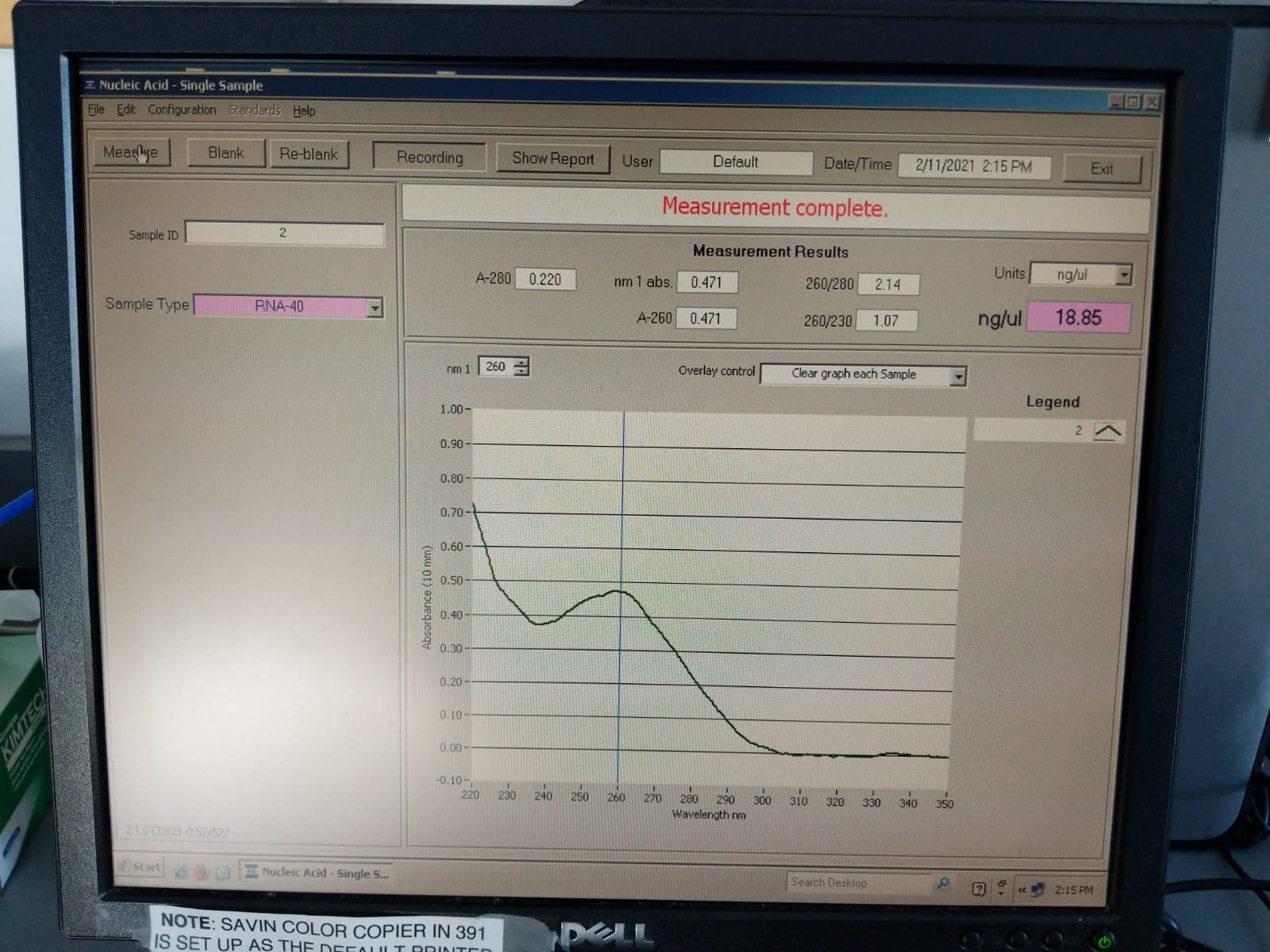
| 2 | 20210131 | M-201 | 2019-12-04 | Bleach | 2.14 | 1.07 | 18.85 |
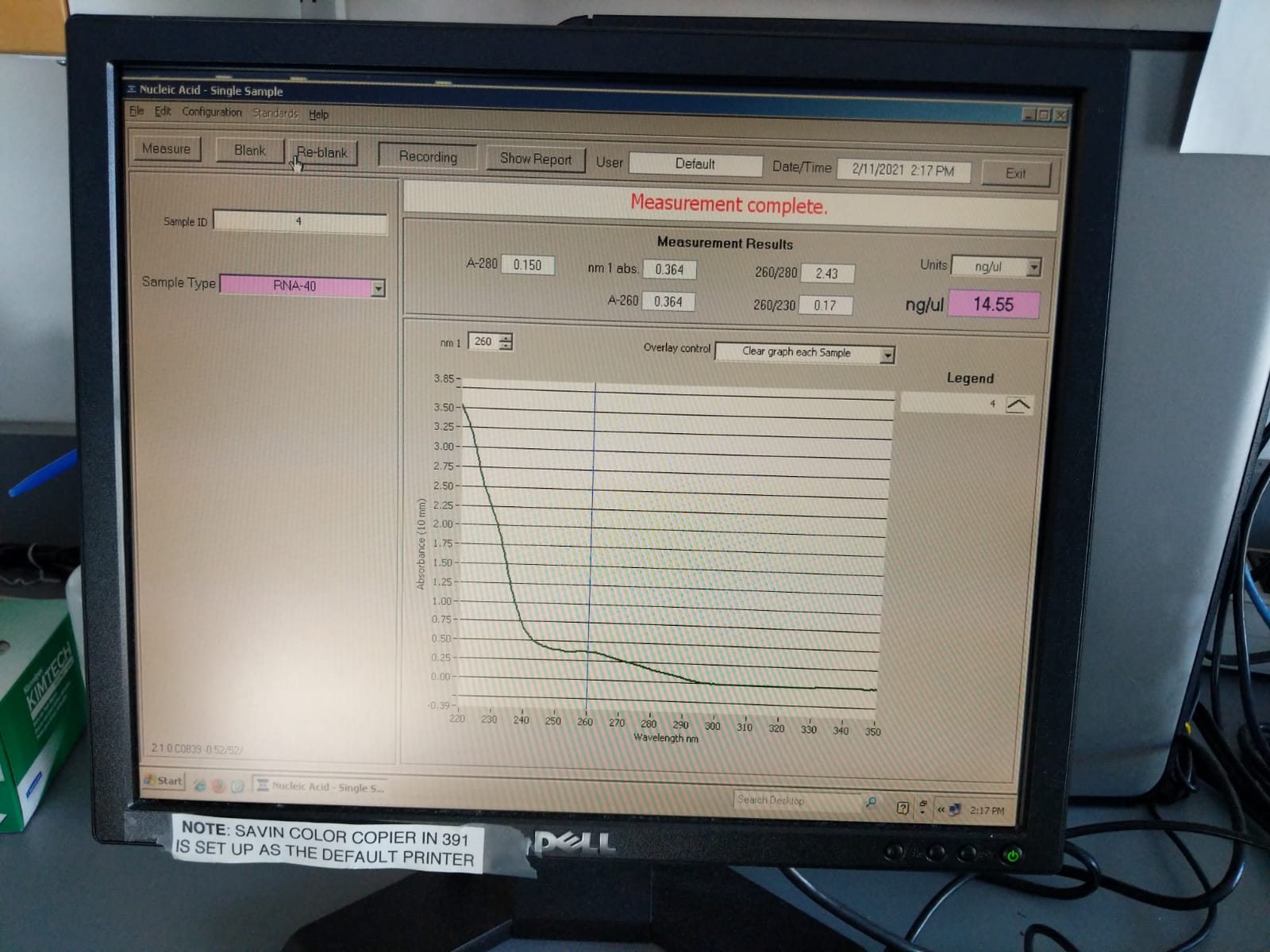
| 4 | 20210131 | M-11 | 2019-12-04 | Bleach | 2.43 | 0.17 | 14.55 |
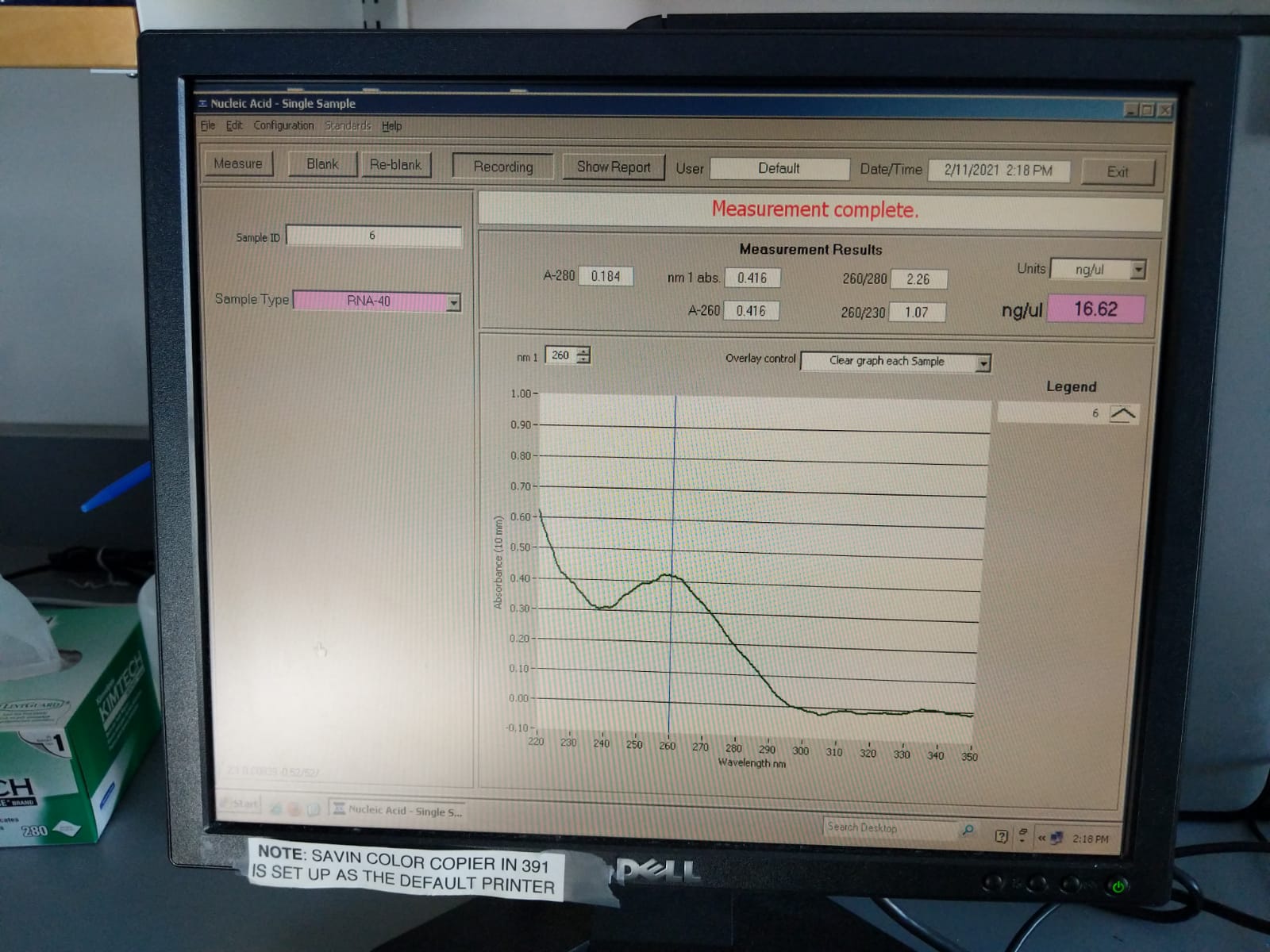
| 6 | 20210131 | M-202 | 2019-12-04 | Non-Bleach | 2.26 | 1.07 | 16.62 |
Extraction ID 2 (M-201):
Extraction ID 4 (M-11):
Extraction ID 6 (M-202):
20210225 Extractions
We are trying to extraction 10/30/2019 instead of 10/2/2019 because in the 20210210 extractions, there was fair quality RNA that we might be able to work with.
Qubit Standards:
DNA BR St1: 180.23 St2: 19,486.52
RNA HS St1: 46.26 St2: 777.31
| ColonyID | Collection-Date | Bleach- | Extraction-ID | Extraction-Date | DNA reading 1 (ng/uL) | DNA reading 2 (ng/uL) | DNA reading Avg (ng/uL) | RNA reading 1 (ng/uL) | RNA reading 2 (ng/uL) | RNA reading Avg (ng/uL) | RIN | Gel Pass? | Final Extraction for this coral? | Notes |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M-209 | 2019-10-30 | Bleach | 13 | 20210225 | 31.8 | 31.6 | 31.7 | 29.2 | 29.2 | 29.2 | 5 | No | NA | Degraded; but potential here |
| M-4 | 2019-10-30 | Non-Bleach | 14 | 20210225 | 28.2 | 28.2 | 28.2 | 26 | 26 | 26 | 3.6 | No | NA | Degraded |
| M-202 | 2019-10-30 | Non-Bleach | 15 | 20210225 | 18.6 | 18.5 | 18.55 | 34 | 34 | 34 | 5 | No | NA | Degraded; but potential here |
| M-217 | 2019-12-04 | Bleach | 16 | 20210225 | 36.6 | 36.4 | 36.5 | 34 | 34 | 34 | 7.7 | Yes | Yes | NA |
| M-212 | 2019-12-04 | Non-Bleach | 17 | 20210225 | 33 | 32.8 | 32.9 | 30.8 | 30.8 | 30.8 | 8.5 | Yes | Yes | NA |
| M-4 | 2019-12-04 | Non-Bleach | 18 | 20210225 | 31.4 | 31.2 | 31.3 | 16.4 | 16.4 | 16.4 | 8.7 | Yes | Yes | NA |
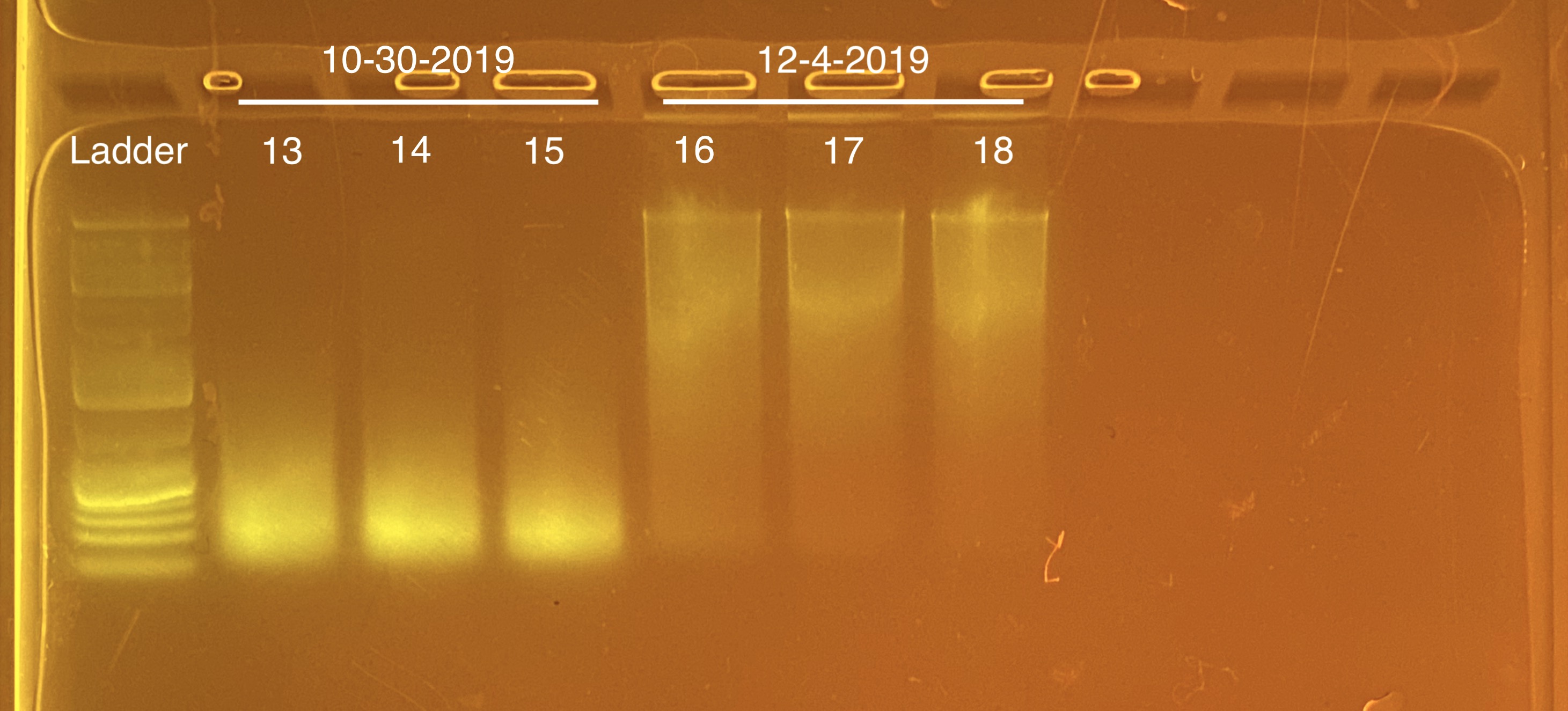
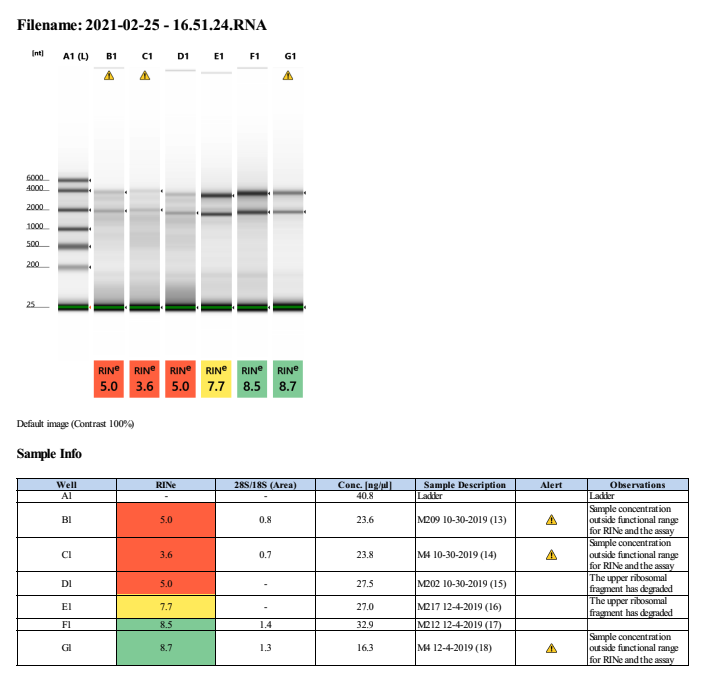
Full tapestation report found here.
20210303 Extractions
Round of 6 fragments from 12-4-2019.
Qubit Standards:
DNA BR St1: 212.09 St2: 19,457.21
RNA HS St1: 46.34 St2: 755.64
| ColonyID | Collection-Date | Bleach- | Extraction-ID | Extraction-Date | DNA reading 1 (ng/uL) | DNA reading 2 (ng/uL) | DNA reading Avg (ng/uL) | RNA reading 1 (ng/uL) | RNA reading 2 (ng/uL) | RNA reading Avg (ng/uL) | RIN | Gel Pass? | Final Extraction for this coral? | Notes |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M-210 | 2019-12-04 | Non-Bleach | 19 | 20210303 | 58.4 | 58.2 | 58.3 | 30 | 30.2 | 30.1 | 7.5 | Yes | Yes | NA |
| M-218 | 2019-12-04 | Non-Bleach | 20 | 20210303 | 64 | 63.8 | 63.9 | 48.8 | 48.8 | 48.8 | 7.4 | Yes | Yes | NA |
| M-20 | 2019-12-04 | Non-Bleach | 21 | 20210303 | 54.6 | 54.4 | 54.5 | 36.8 | 36.8 | 36.8 | 8.1 | Yes | Yes | NA |
| M-209 | 2019-12-04 | Bleach | 22 | 20210303 | 51.2 | 51 | 51.1 | 37.8 | 37.8 | 37.8 | 8.5 | Yes | Yes | NA |
| M-219 | 2019-12-04 | Bleach | 23 | 20210303 | 35.8 | 35.6 | 35.7 | 30 | 30 | 30 | 9.1 | Yes | Yes | NA |
| M-220 | 2019-12-04 | Non-Bleach | 24 | 20210303 | 69 | 68.8 | 68.9 | 44.8 | 44.8 | 44.8 | 9.3 | Yes | Yes | NA |
Gel - extraction samples are the first six: in order of extraction number (19, 20, 21, 22, 23, 24)
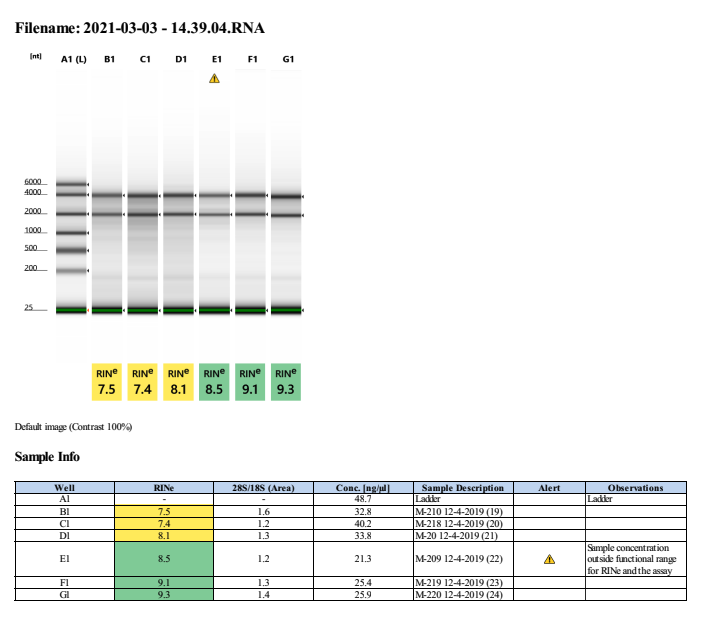
Full tapestation report found here.
20210305 Extractions
Qubit Standards:
DNA BR St1: 163.81 St2: 17,152.69
RNA HS St1: 48.13 St2: 795.17
| ColonyID | Collection-Date | Bleach- | Extraction-ID | Extraction-Date | DNA reading 1 (ng/uL) | DNA reading 2 (ng/uL) | DNA reading Avg (ng/uL) | RNA reading 1 (ng/uL) | RNA reading 2 (ng/uL) | RNA reading Avg (ng/uL) | RIN | Gel Pass? | Final Extraction for this coral? | Notes |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M-222 | 2019-12-04 | Non-Bleach | 26 | 20210305 | 46.2 | 45.8 | 46 | 28.8 | 28.6 | 28.7 | 6.8 | |||
| M-3 | 2019-12-04 | Bleach | 27 | 20210305 | 21 | 20.8 | 20.9 | 12.2 | 12.3 | 12.25 | NA | No | RNA and RIN score too low | |
| M-221 | 2019-12-04 | Bleach | 28 | 20210305 | 30.4 | 30.8 | 30.6 | 30.8 | 30.6 | 30.7 | 8.6 | |||
| M-19 | 2019-12-04 | Bleach | 29 | 20210305 | 64.2 | 63.4 | 63.8 | 51.6 | 51.2 | 51.4 | 7.3 | |||
| M-204 | 2019-12-04 | Non-Bleach | 30 | 20210305 | 37.6 | 37.4 | 37.5 | 31.8 | 31.6 | 31.7 | 8.2 | |||
| M-203 | 2019-12-04 | Bleach | 31 | 20210305 | 21.2 | 21.2 | 21.2 | 23 | 23 | 23 | 8.7 | |||
| M-211 | 2019-12-04 | Bleach | 32 | 20210305 | 27 | 26.8 | 26.9 | 18.7 | 18.7 | 18.7 | 9.2 |
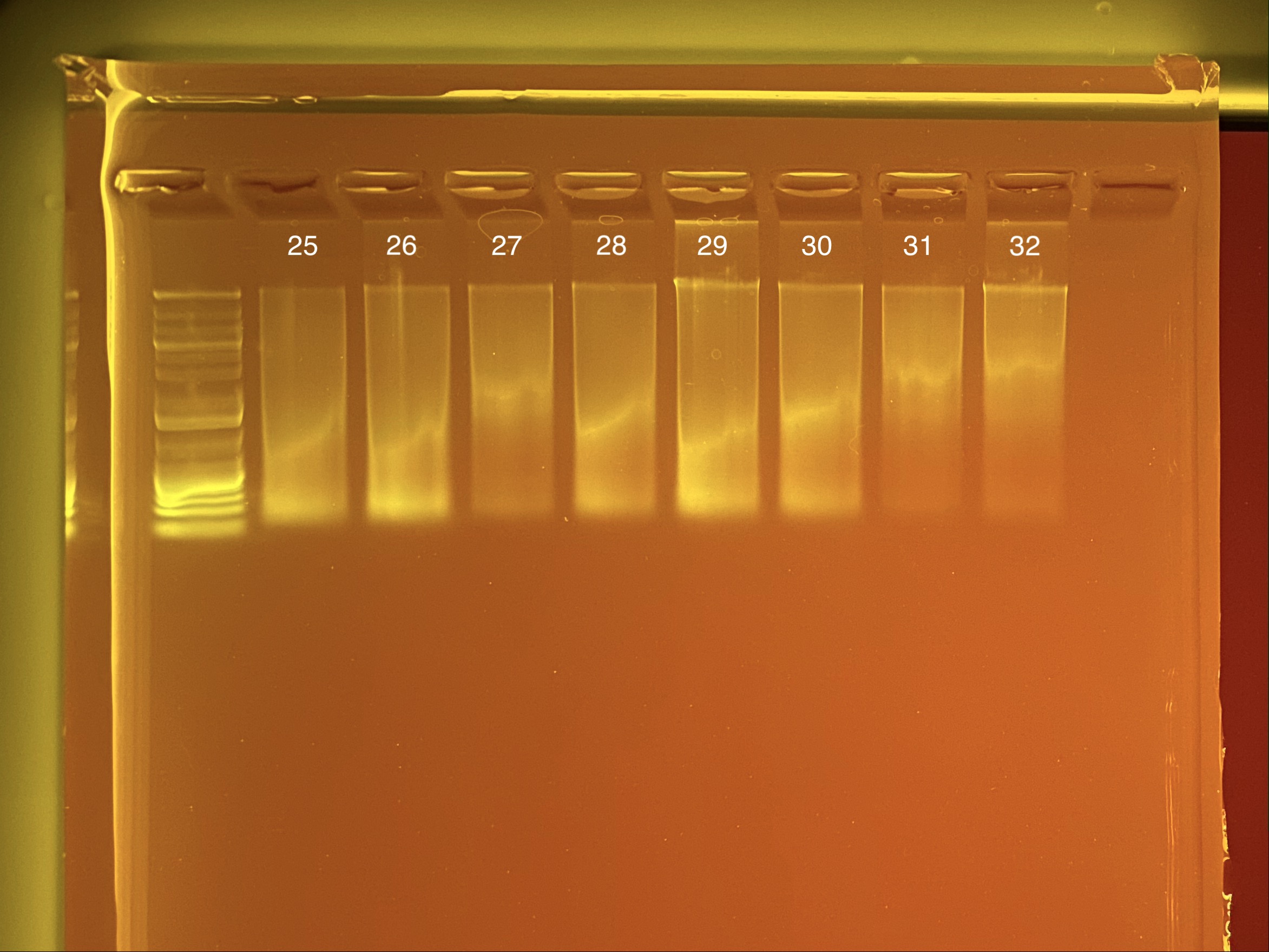
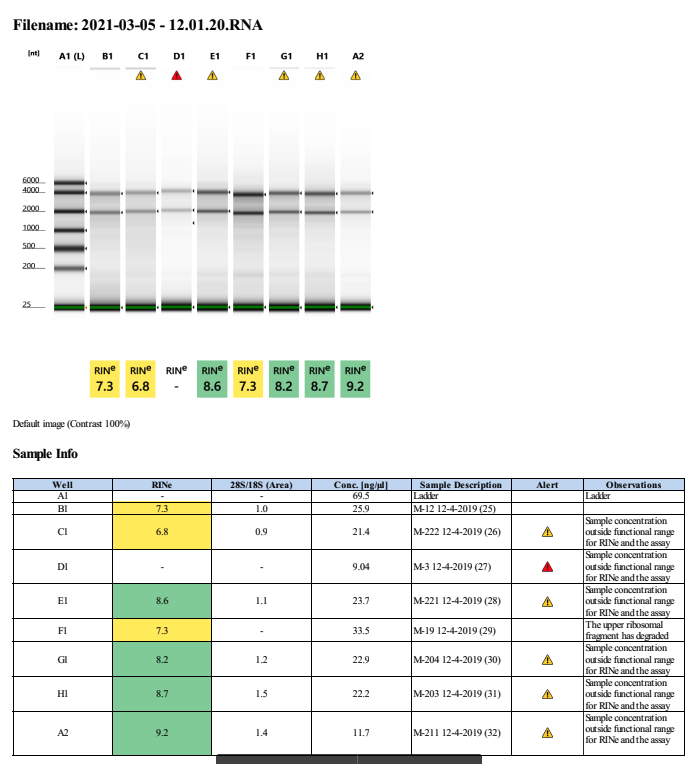
Extraction ID 27 was re-done on the tapestation but the same result occured. Need to redo this coral.
Full tapestation report found here.
List to re-do: M-3, M-210, M-218
20210316 Extractions
Qubit Standards:
DNA BR St1: 171.22 St2: 19,153.81
RNA HS St1: 47.58 St2: 836.09
| ColonyID | Collection-Date | Bleach- | Extraction-ID | Extraction-Date | DNA reading 1 (ng/uL) | DNA reading 2 (ng/uL) | DNA reading Avg (ng/uL) | RNA reading 1 (ng/uL) | RNA reading 2 (ng/uL) | RNA reading Avg (ng/uL) | RIN | Gel Pass? | Final Extraction for this coral? | Notes |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M-218 | 2019-12-04 | Non-Bleach | 33 | 20210316 | 89.6 | 89.2 | 89.4 | 56.8 | 56.8 | 56.8 | 8.4 | Yes | Yes | |
| M-210 | 2019-12-04 | Non-Bleach | 34 | 20210316 | 37.3 | 37 | 37.15 | 18.6 | 18.6 | 18.6 | 8.2 | Yes | Yes | |
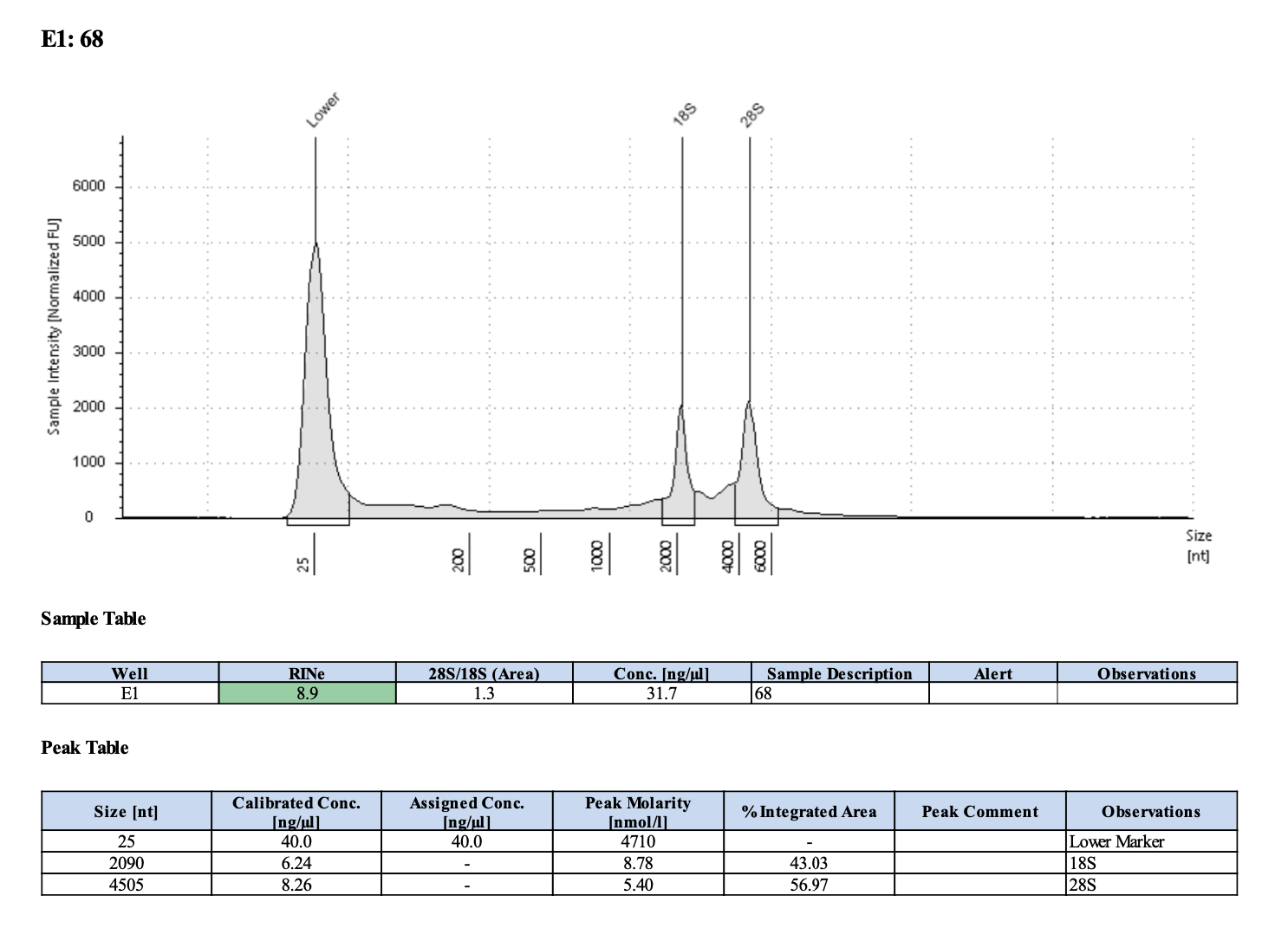
| M-3 | 2019-12-04 | Bleach | 35 | 20210316 | 32.2 | 32 | 32.1 | 17.3 | 17.2 | 17.25 | 8.9 | Yes | Yes |
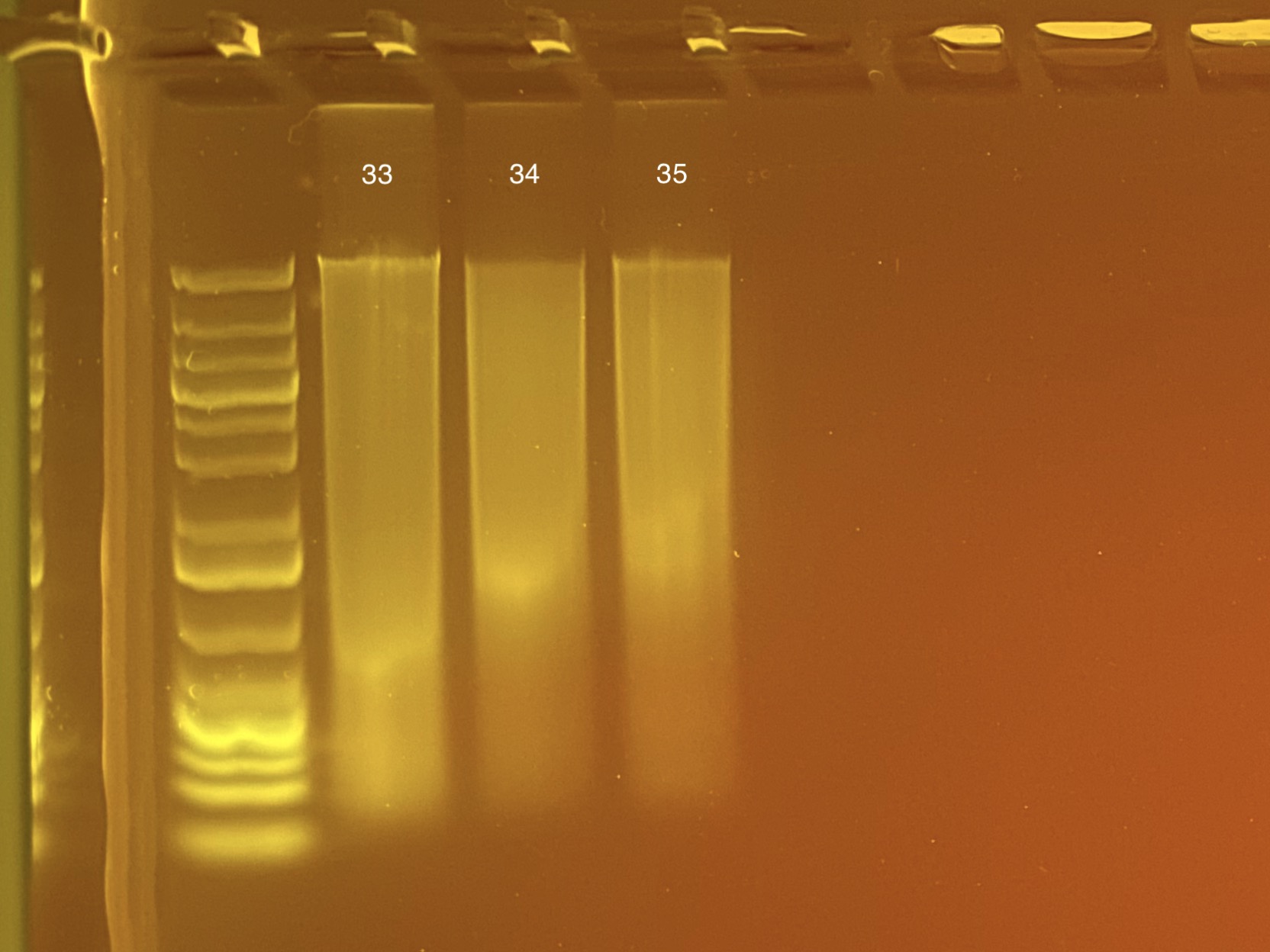
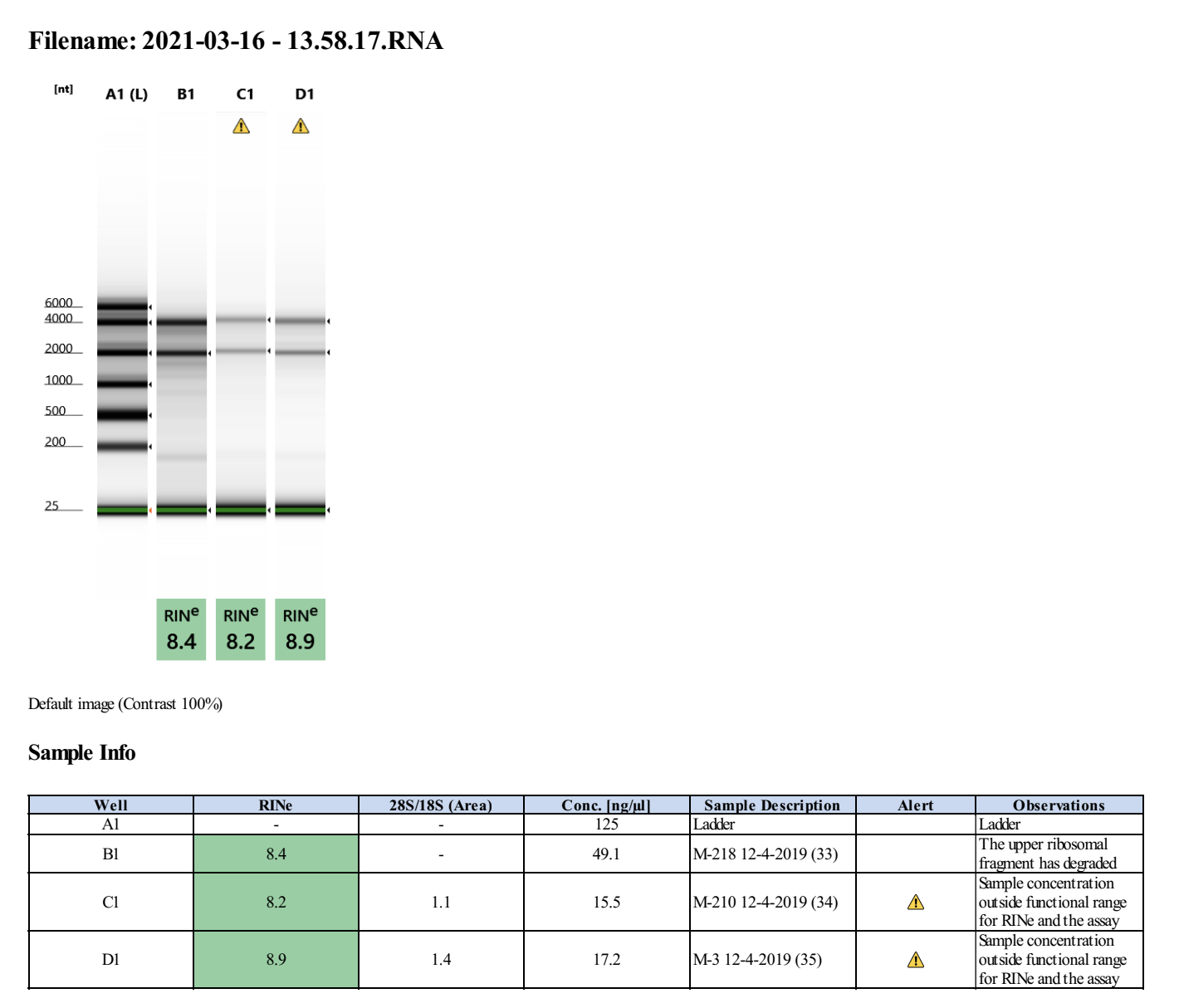
Full tapestation report found here.
July 2019 Extractions
20210430 Extractions
Qubit Standards:
DNA BR St1: 132.73 St2: 25,435.25
RNA HS St1: 48.59 St2: 941.24
| ColonyID | Collection-Date | Bleach- | Extraction-ID | Extraction-Date | DNA reading 1 (ng/uL) | DNA reading 2 (ng/uL) | DNA reading Avg (ng/uL) | RNA reading 1 (ng/uL) | RNA reading 2 (ng/uL) | RNA reading Avg (ng/uL) | RIN | Gel Pass? | Final Extraction for this coral? | Notes |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
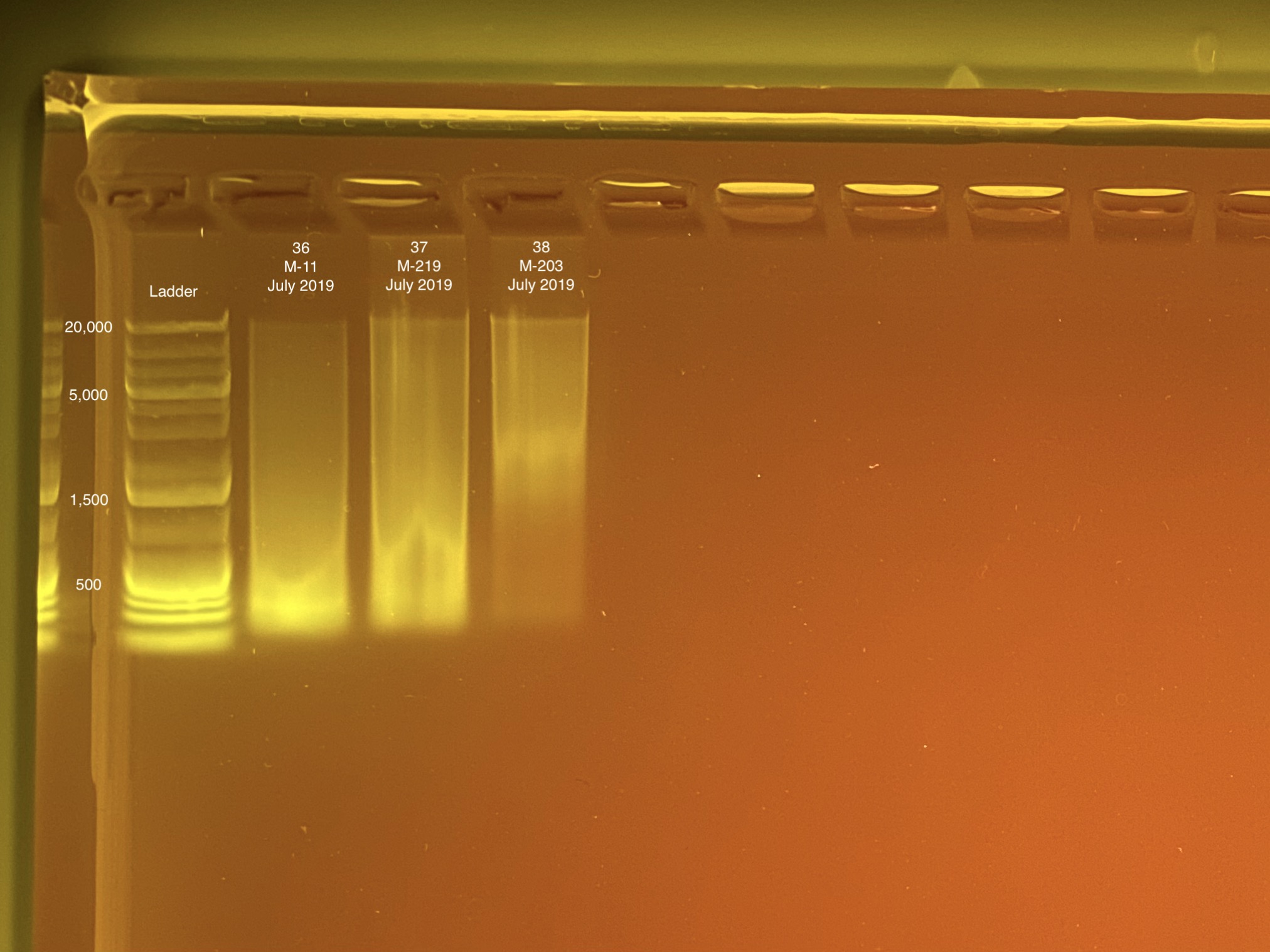
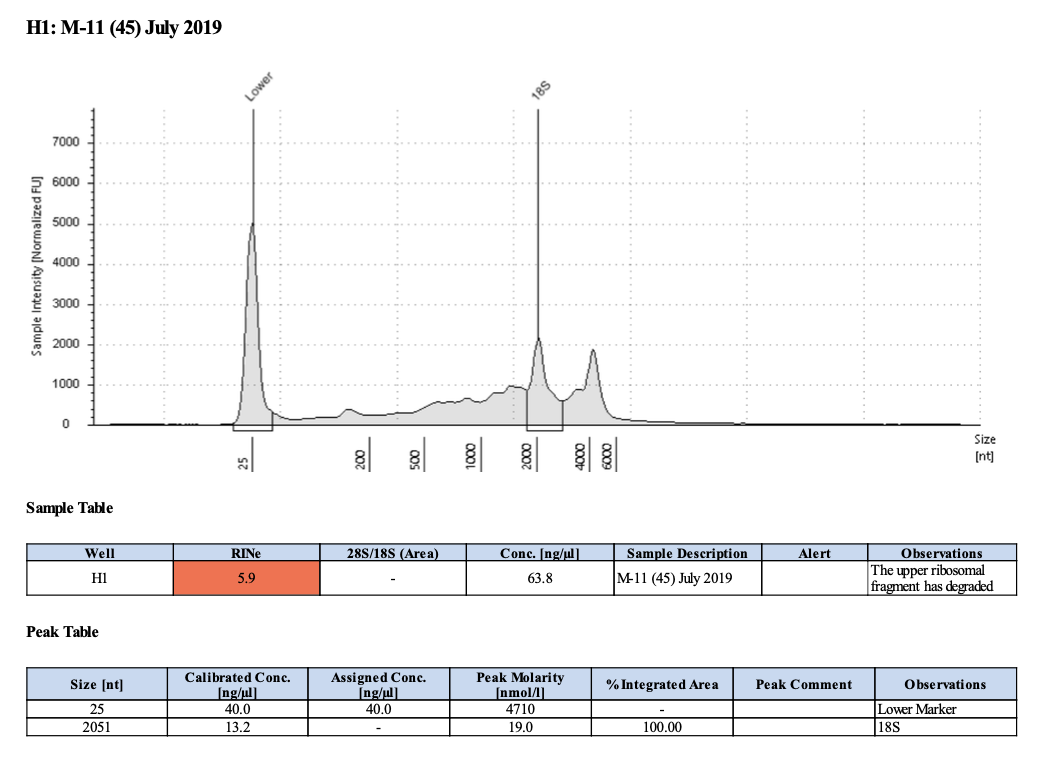
| M-11 | 2019-07-20 | Bleach | 36 | 20210430 | 28.4 | 28.2 | 28.3 | 64.8 | 64.4 | 64.6 | 5.6 | No | No | RIN too low |
| M-219 | 2019-07-20 | Bleach | 37 | 20210430 | 34 | 33.8 | 33.9 | 32.8 | 32.8 | 32.8 | 8 | No | TBD | maybe re-do this one b/c the DNA quality is low |
| M-203 | 2019-07-20 | Bleach | 38 | 20210430 | 15.2 | 15 | 15.1 | 13.5 | 13.6 | 13.55 | 8.4 | Yes | Yes | NA |
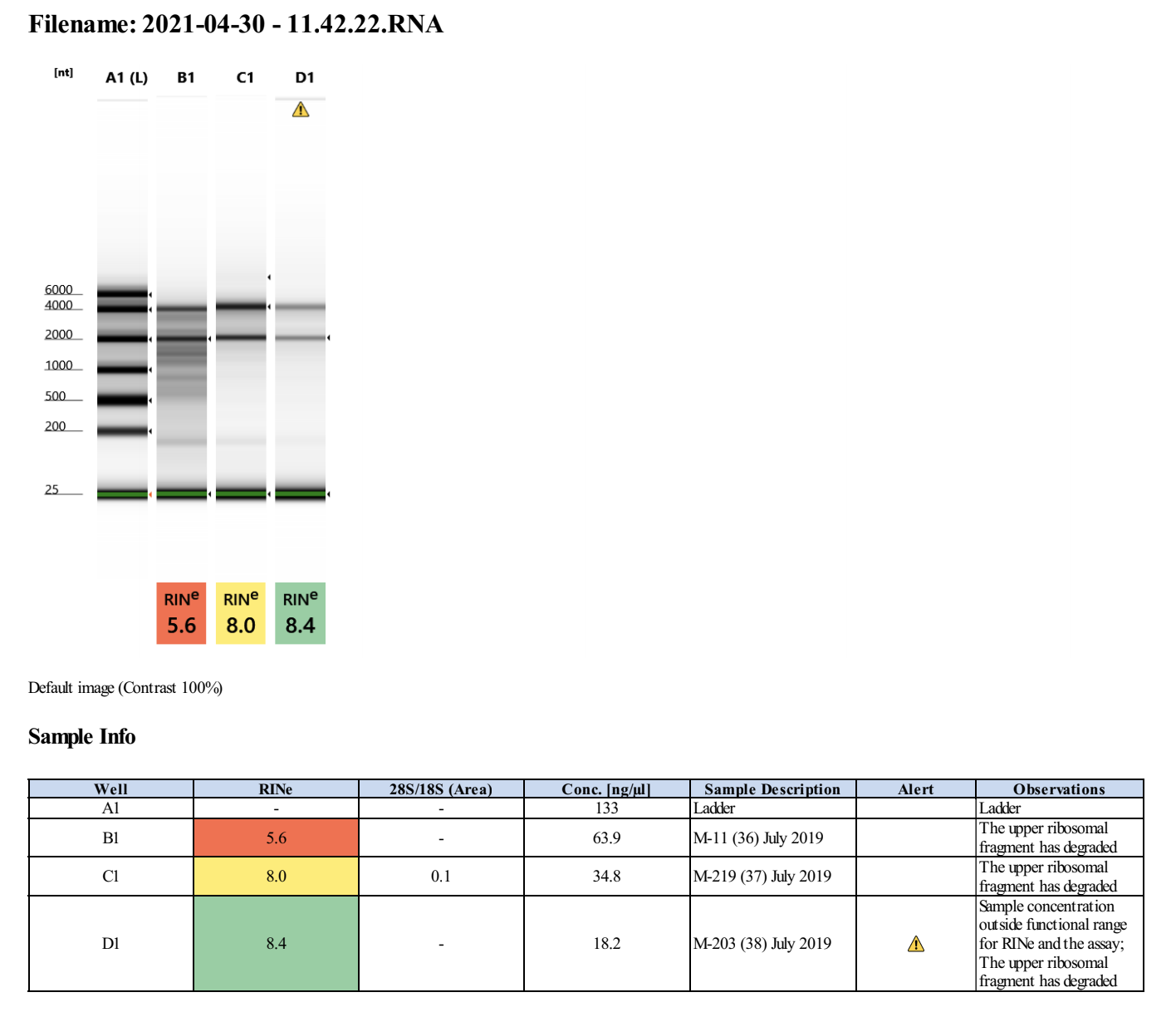
Full tapestation report found here.
Corals to re-do:
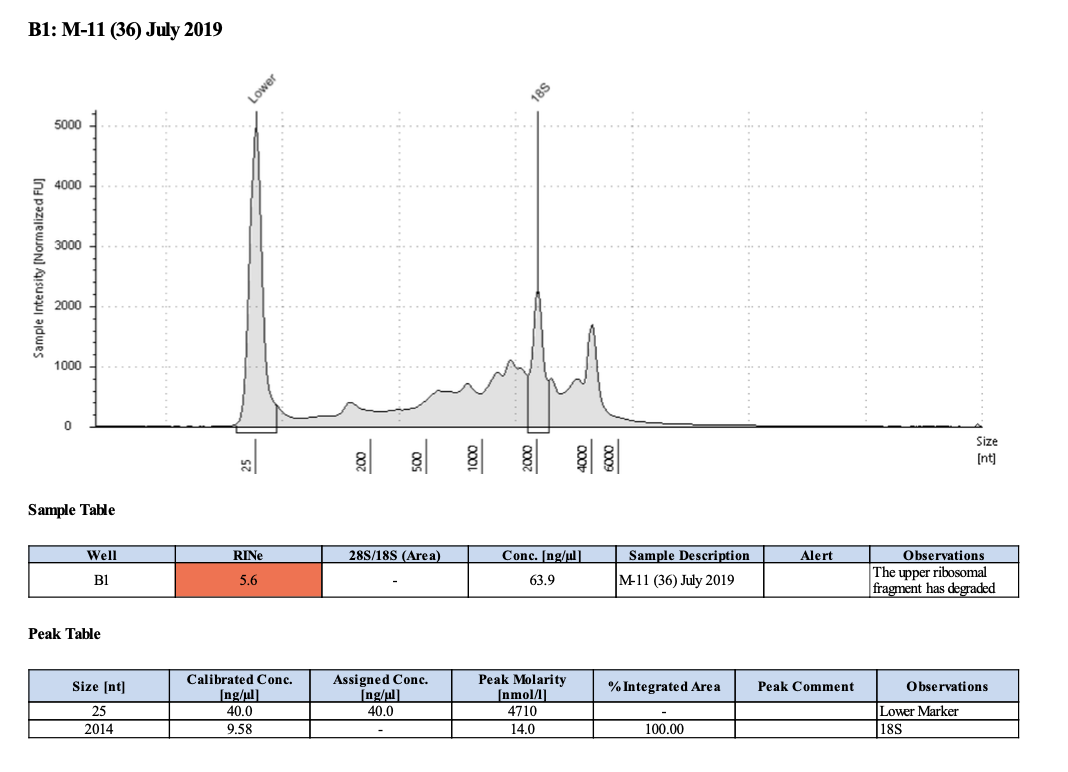
20210503 Extractions
Qubit Standards:
DNA BR St1: 192.69 St2: 21,389.02
RNA HS St1: 48.65 St2: 827.45
| ColonyID | Collection-Date | Bleach- | Extraction-ID | Extraction-Date | DNA reading 1 (ng/uL) | DNA reading 2 (ng/uL) | DNA reading Avg (ng/uL) | RNA reading 1 (ng/uL) | RNA reading 2 (ng/uL) | RNA reading Avg (ng/uL) | RIN | Gel Pass? | Final Extraction for this coral? |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M-212 | 2019-07-20 | Non-Bleach | 39 | 20210503 | 105 | 103 | 104 | 73.6 | 72.8 | 73.2 | 6.2 | No | TBD |
| M-209 | 2019-07-20 | Bleach | 40 | 20210503 | 40.2 | 39.8 | 40 | 16.1 | 16 | 16.05 | 8.2 | No | TBD |
| M-3 | 2019-07-20 | Bleach | 41 | 20210503 | 28.8 | 28.4 | 28.6 | 11.2 | 11.2 | 11.2 | 6.5 | No | TBD |
| M-222 | 2019-07-20 | Non-Bleach | 42 | 20210503 | 80.6 | 79.8 | 80.2 | 36.8 | 37 | 36.9 | 7.1 | No | TBD |
| M-210 | 2019-07-20 | Non-Bleach | 43 | 20210503 | 47.6 | 47.2 | 47.4 | 26 | 26 | 26 | 7.2 | No | TBD |
| M-201 | 2019-07-20 | Bleach | 44 | 20210503 | 37.2 | 37 | 37.1 | 12.8 | 12.7 | 12.75 | 8.1 | No | TBD |
| M-11 | 2019-07-20 | Bleach | 45 | 20210503 | 73.8 | 73.2 | 73.5 | 76.6 | 76.2 | 76.4 | 5.9 | No | TBD |
| M-202 | 2019-07-20 | Non-Bleach | 46 | 20210503 | 89 | 88.2 | 88.6 | 71.8 | 71.4 | 71.6 | 6.8 | No | TBD |
| M-217 | 2019-07-20 | Bleach | 47 | 20210503 | 80.4 | 79.6 | 80 | 46 | 45.8 | 45.9 | 9 | No | TBD |
| M-219 | 2019-07-20 | Bleach | 48 | 20210503 | 149 | 148 | 148.5 | 96.9 | 95.8 | 96.35 | 6.9 | No | TBD |
| M-20 | 2019-07-20 | Non-Bleach | 49 | 20210503 | 17.7 | 17.5 | 17.6 | 21.4 | 21.4 | 21.4 | 3.8 | No | TBD |
| M-204 | 2019-07-20 | Non-Bleach | 50 | 20210503 | 69.6 | 69 | 69.3 | 44.4 | 44.2 | 44.3 | 6 | No | TBD |
| M-221 | 2019-07-20 | Bleach | 51 | 20210503 | 40.4 | 40.2 | 40.3 | 24.6 | 24.6 | 24.6 | 7.9 | No | TBD |
| M-211 | 2019-07-20 | Bleach | 52 | 20210503 | 75 | 74.4 | 74.7 | 40.2 | 40 | 40.1 | 6.7 | No | TBD |
| M-220 | 2019-07-20 | Non-Bleach | 53 | 20210503 | 21.4 | 21.2 | 21.3 | 10.2 | 10.3 | 10.25 | NA | Yes | TBD |
| M-4 | 2019-07-20 | Non-Bleach | 54 | 20210503 | 112 | 111 | 111.5 | 72.4 | 72 | 72.2 | 7.3 | No | TBD |
| M-12 | 2019-07-20 | Non-Bleach | 55 | 20210503 | 72.8 | 72 | 72.4 | 55.6 | 55.4 | 55.5 | 7.3 | No | TBD |
| M-218 | 2019-07-20 | Non-Bleach | 56 | 20210503 | 82.2 | 81.4 | 81.8 | 60.4 | 60.4 | 60.4 | 6.6 | No | TBD |
| M-19 | 2019-07-20 | Bleach | 57 | 20210503 | 65.6 | 65.2 | 65.4 | 34.8 | 34.8 | 34.8 | 8.1 | No | TBD |
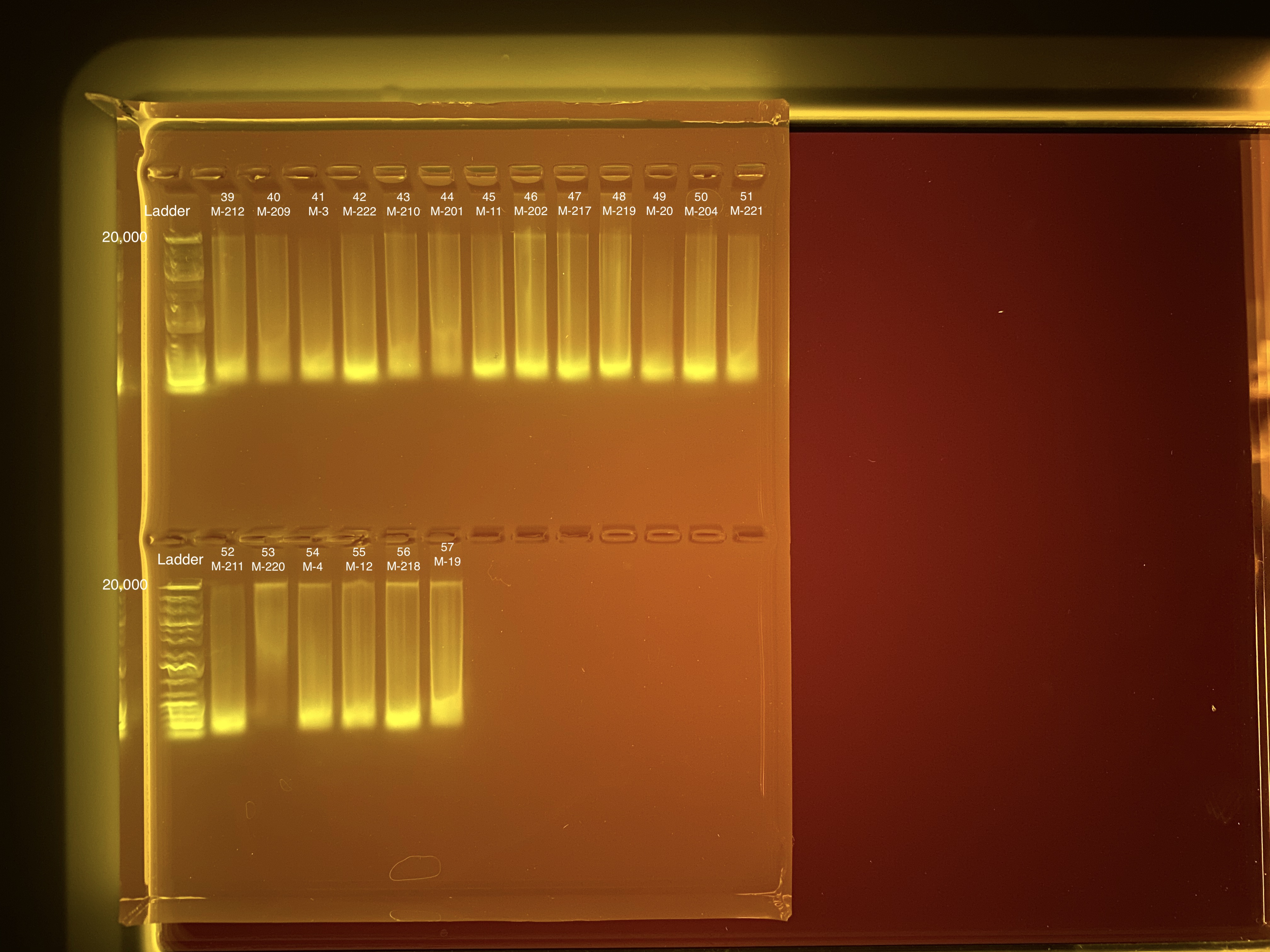
Full tapestation report found here and here.
20210517 Extractions
Qubit Standards:
DNA BR St1: 171.33 St2: 17,943.96
RNA HS St1: 44.48 St2: 717.72
| ColonyID | Collection-Date | Bleach- | Extraction-ID | Extraction-Date | DNA reading 1 (ng/uL) | DNA reading 2 (ng/uL) | DNA reading Avg (ng/uL) | RNA reading 1 (ng/uL) | RNA reading 2 (ng/uL) | RNA reading Avg (ng/uL) | RIN | Gel Pass? | Final Extraction for this coral? | Notes |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M-11 | 2019-07-20 | Bleach | 58 | 20210517 | 55.4 | 55.4 | 55.4 | 60.8 | 60.8 | 60.8 | 5.4 | Enough | No | This isn’t much better than the first two |
| M-20 | 2019-07-20 | Non-Bleach | 59 | 20210517 | 18.4 | 18.3 | 18.35 | 20 | 19.9 | 19.95 | 5.1 | Enough | Yes |
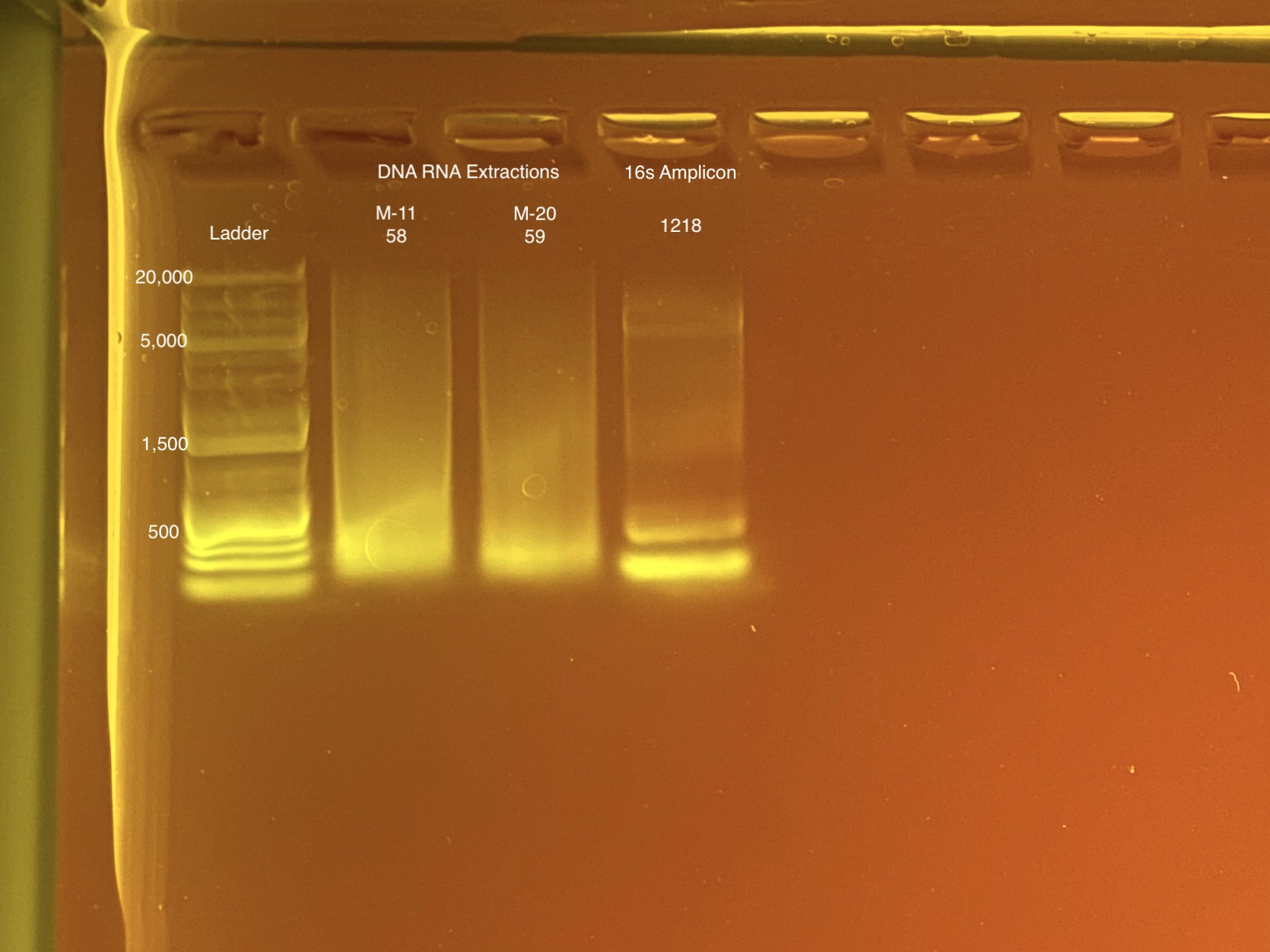
Full tapestation report found here
RNA Quality



















TagSeq and RNASeq
We sequenced the RNA with TagSeq but this failed so we want to ship the RNA to Genewiz for RNASeq. We don’t have enough RNA (1.2 ug needed) from 10/40 of the fragments to send. I re-extracted those 10 fragments and will send that RNA.
Re-extractions for the above 10
| Coral ID | Collection Date | Re-extraction ID | Re-extraction Date | DNA reading 1 (ng/uL) | DNA reading 2 (ng/uL) | DNA reading Avg (ng/uL) | RNA reading 1 (ng/uL) | RNA reading 2 (ng/uL) | RNA reading Avg (ng/uL) | RIN | Gel Pass? | Final Extraction for this coral? | Notes |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
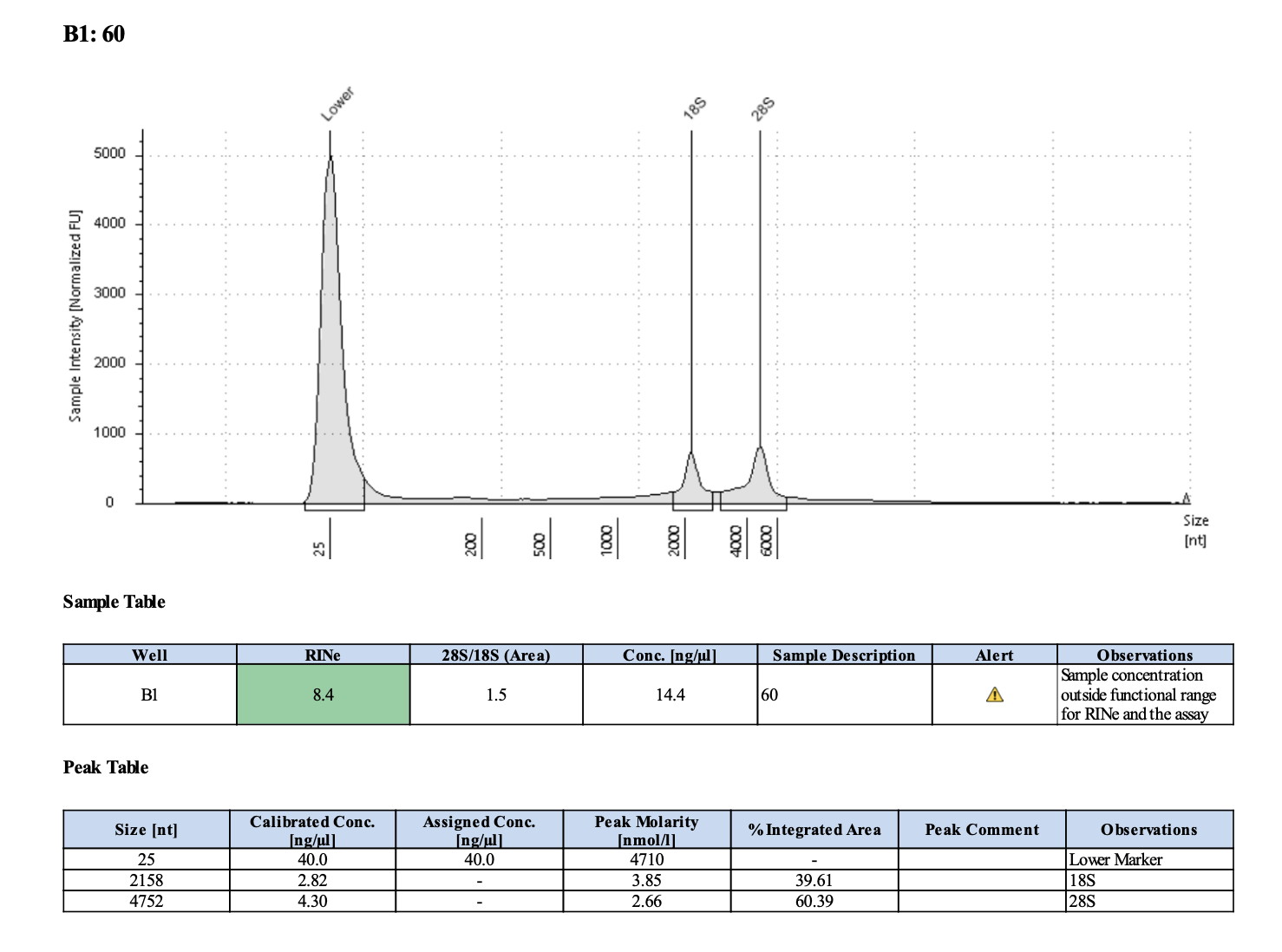
| M-3 | 2019-12-04 | 60 | 20211028 | 28.4 | 28.2 | 28.3 | 13.6 | 13.7 | 13.65 | 8.4 | Yes | Yes | |
| M-211 | 2019-12-04 | 61 | 20211028 | 35.4 | 35.2 | 35.3 | 18.2 | 18.2 | 18.2 | 8.7 | Yes | Yes | |
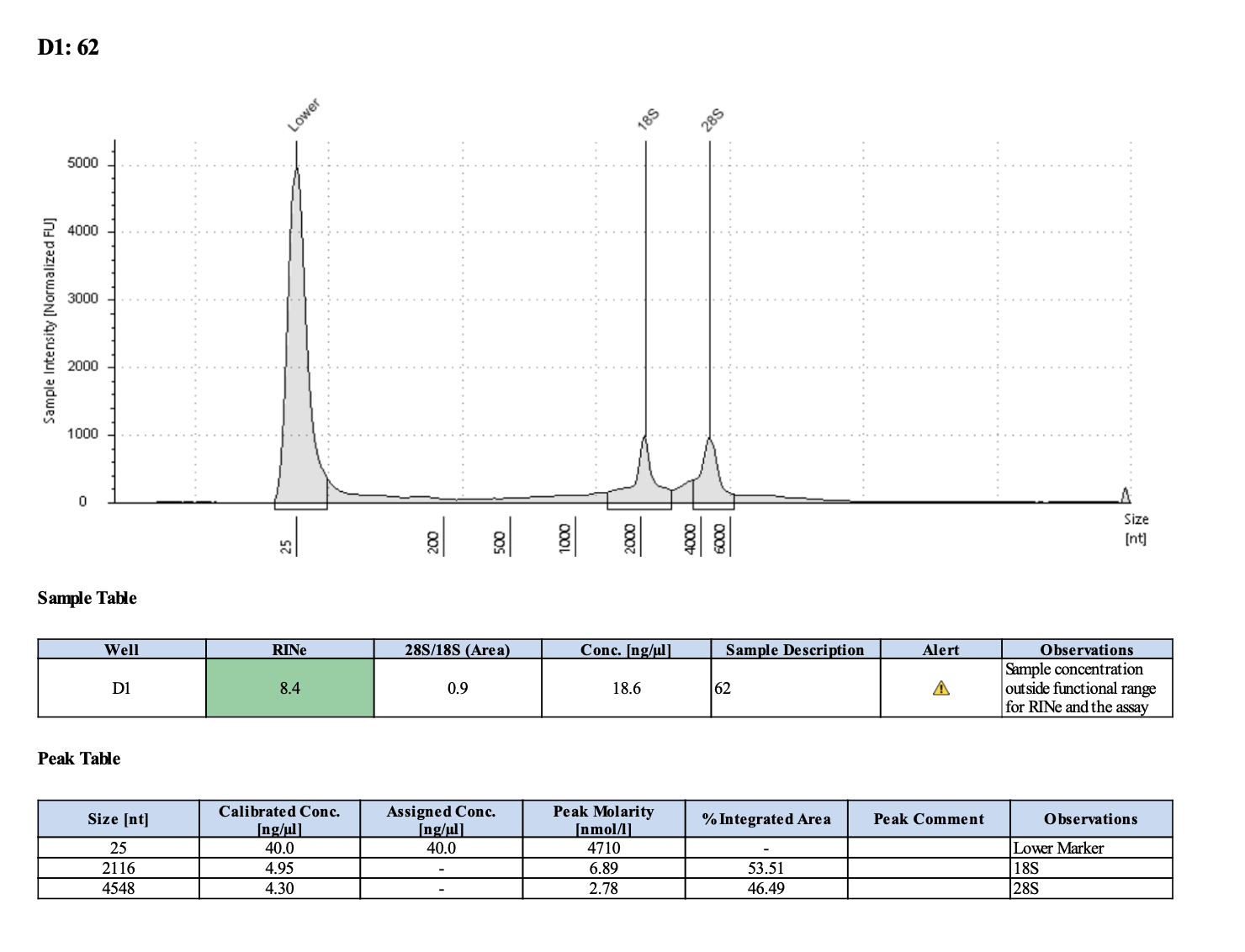
| M-201 | 2019-07-20 | 62 | 20211028 | 52.4 | 52.2 | 52.3 | 19.5 | 19.5 | 19.5 | 8.4 | Enough | Yes | DNA is much more degraded than the December fragments |
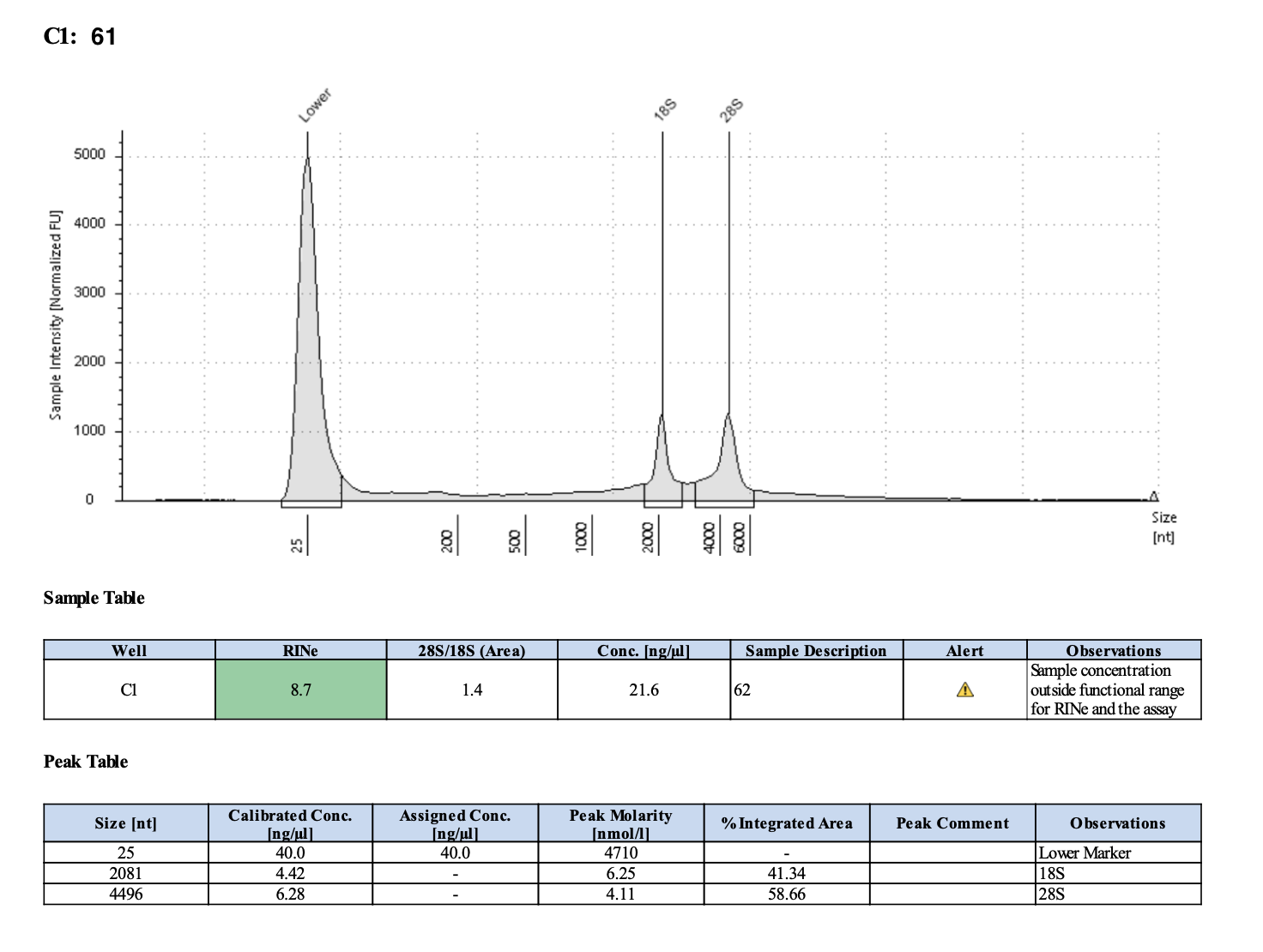
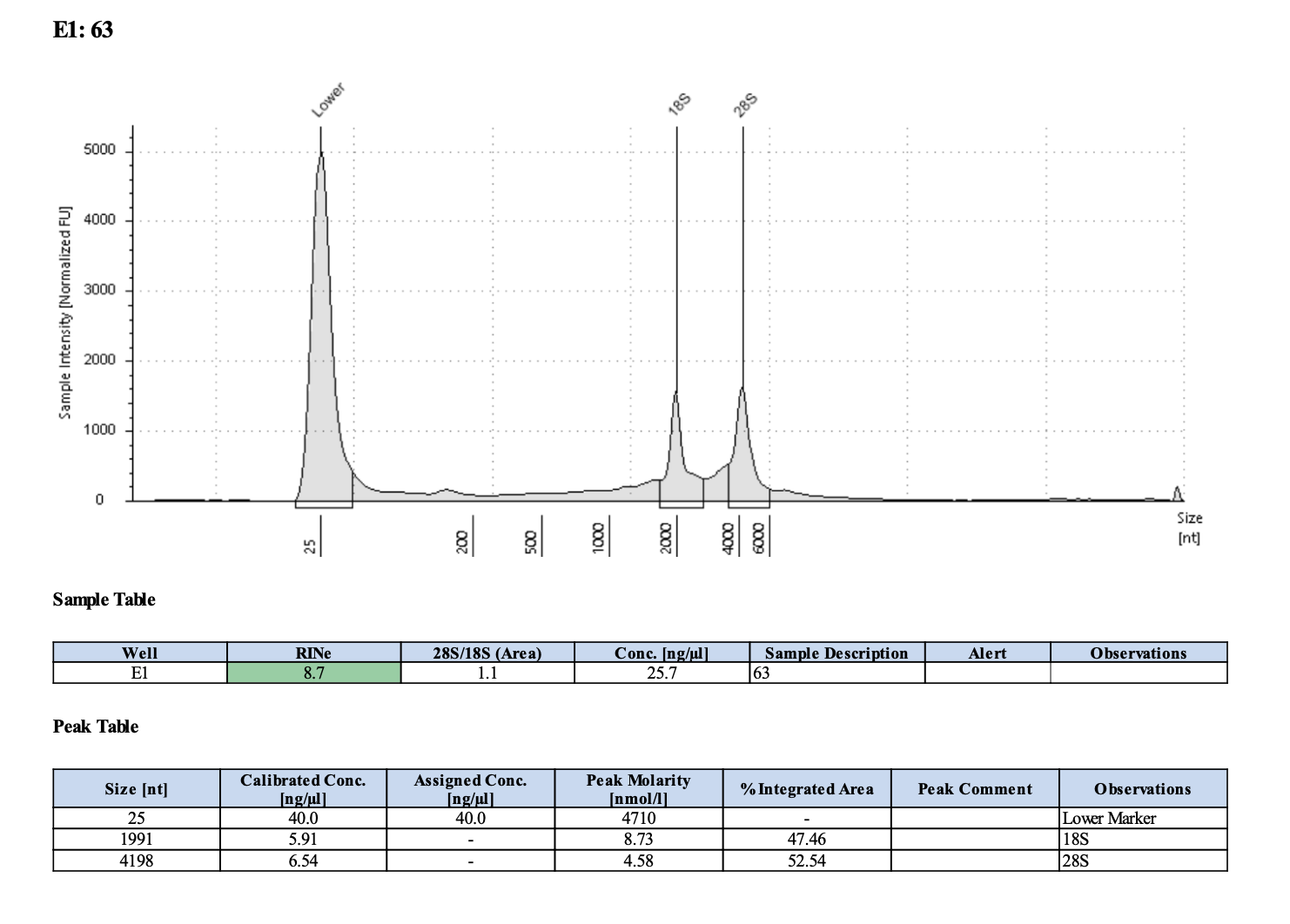
| M-203 | 2019-07-20 | 63 | 20211028 | 55.8 | 55.6 | 55.7 | 32 | 32 | 32 | 8.7 | Enough | Yes | DNA is much more degraded than the December fragments |
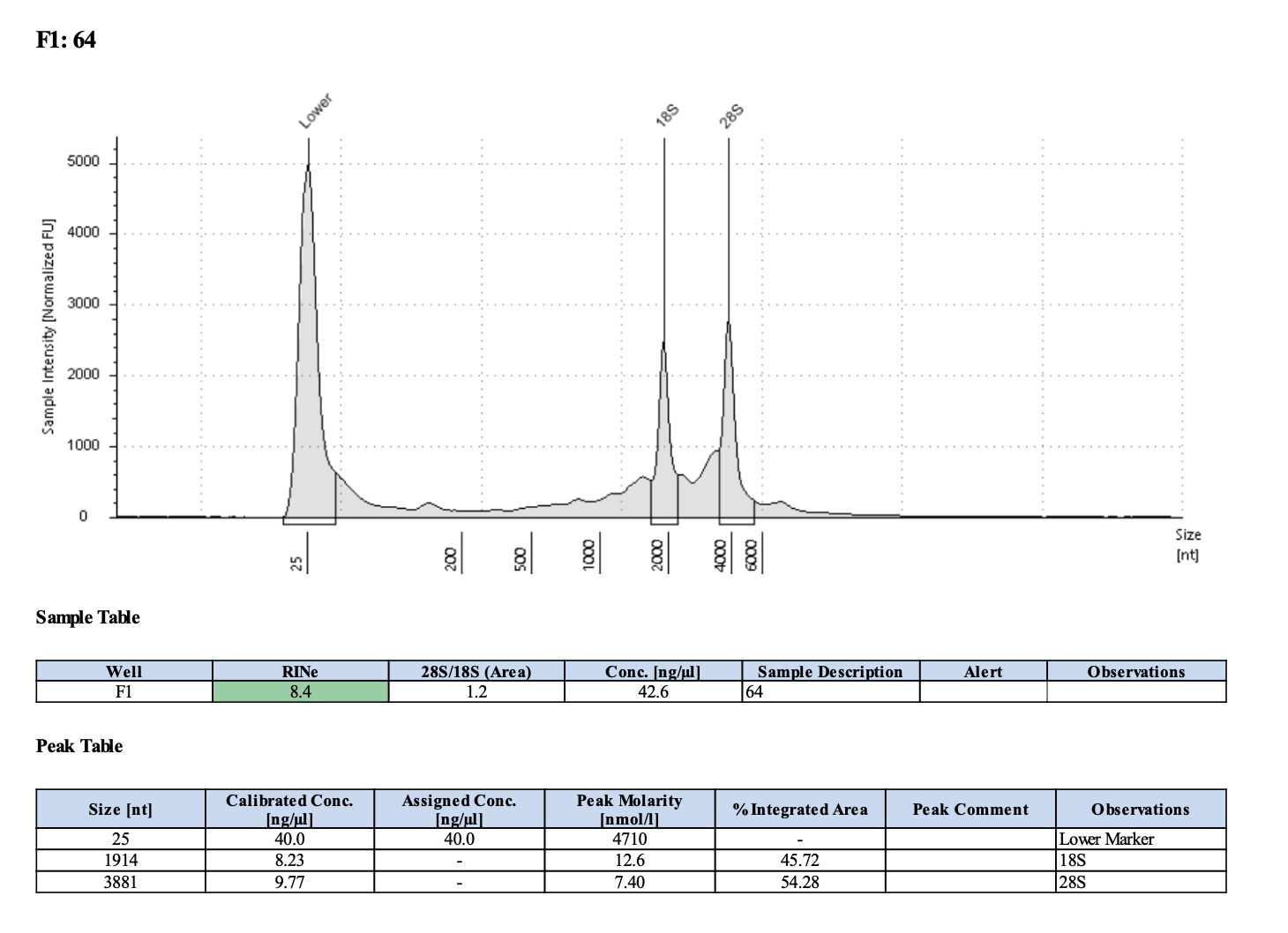
| M-220 | 2019-07-20 | 64 | 20211028 | 101 | 101 | 101 | 49.4 | 49.4 | 49.4 | 8.4 | Enough | Yes | DNA is much more degraded than the December fragments |
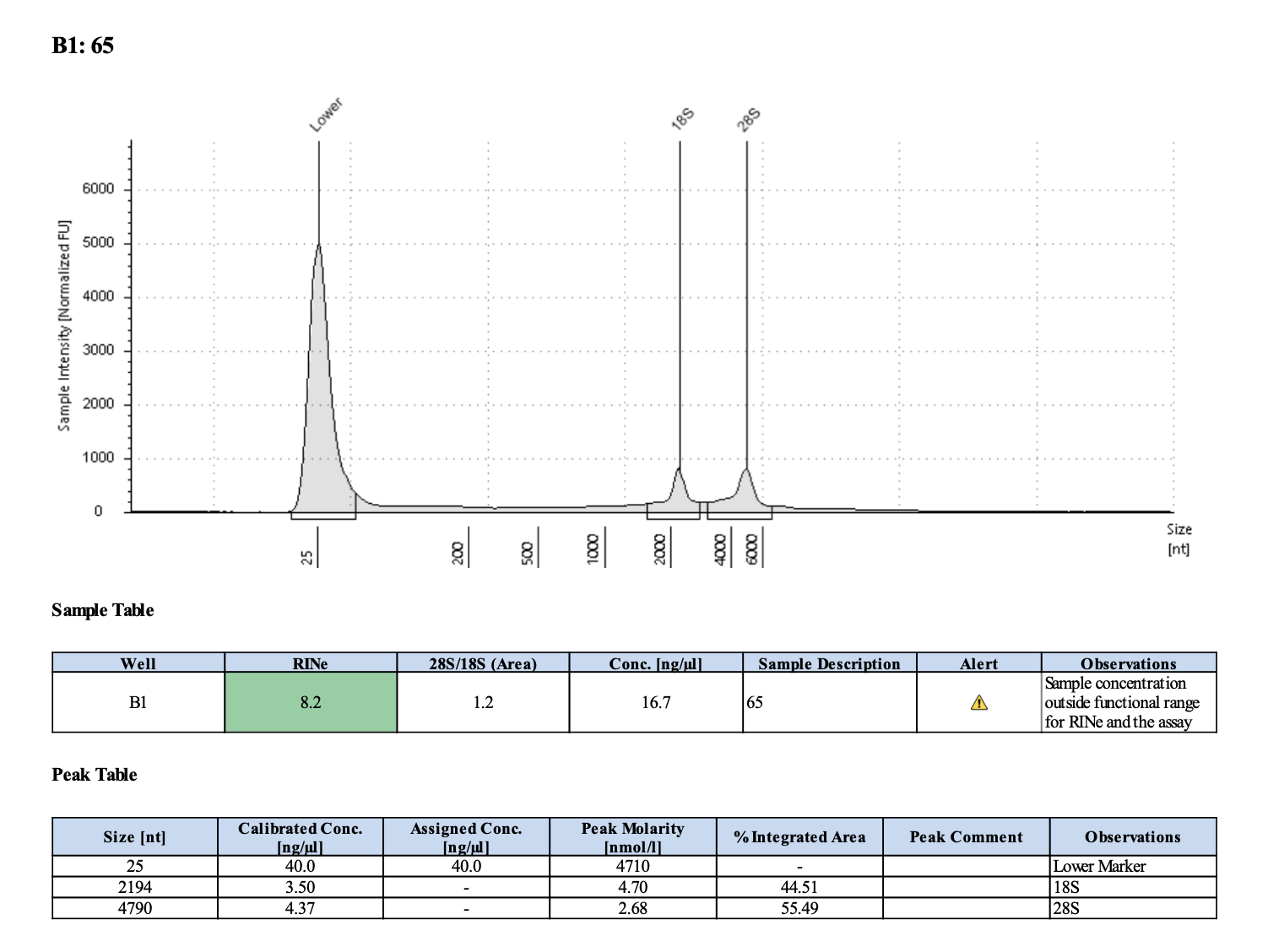
| M-4 | 2019-12-04 | 65 | 20211029 | 30 | 29.8 | 29.9 | 19.2 | 19.1 | 19.15 | 8.2 | Yes | Yes | |
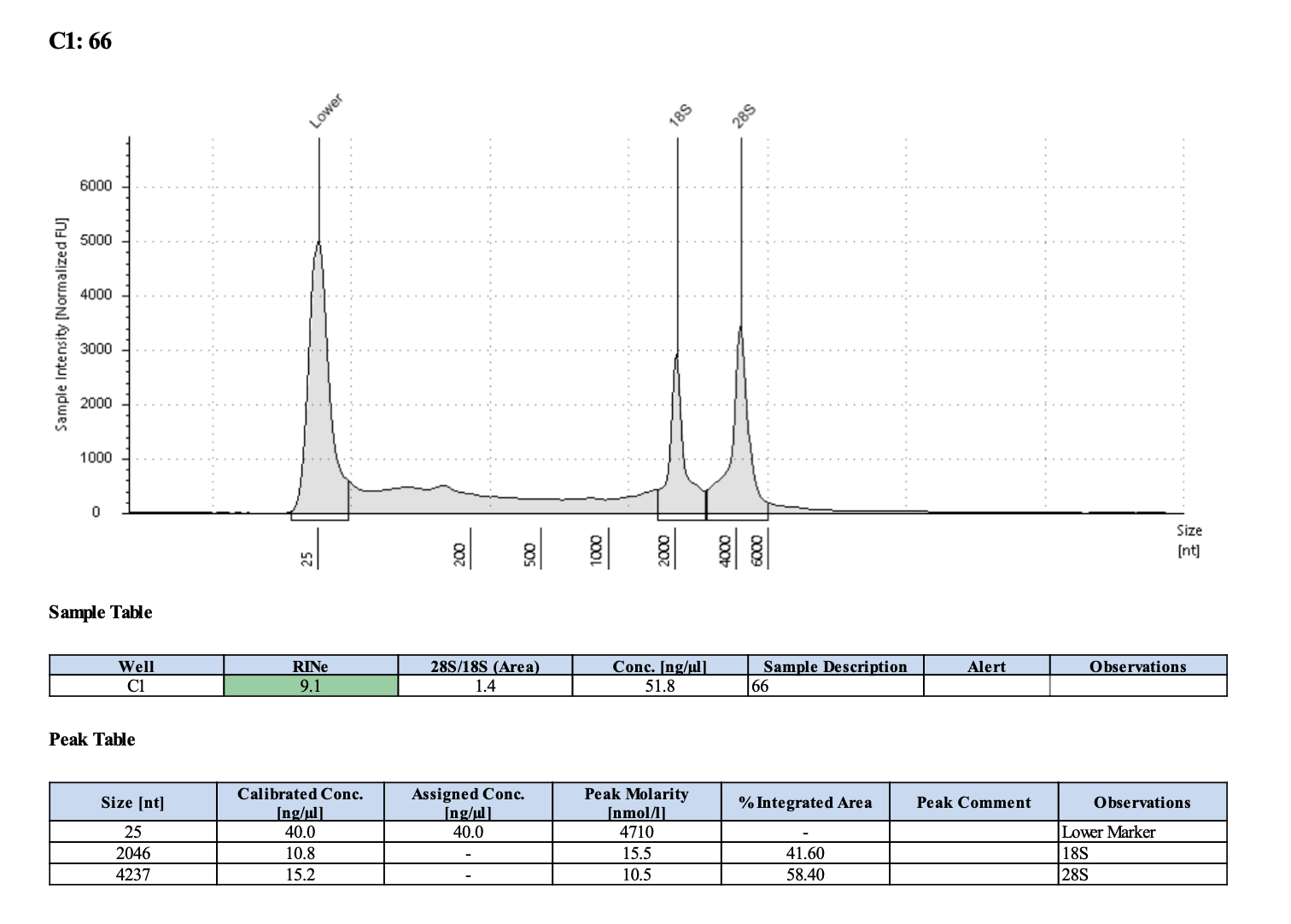
| M-210 | 2019-12-04 | 66 | 20211029 | 46.8 | 46.6 | 46.7 | 48.6 | 48.6 | 48.6 | 9.1 | Yes | Yes | |
| M-209 | 2019-07-20 | 68 | 20211029 | 46.4 | 46.4 | 46.4 | 31.8 | 31.8 | 31.8 | 5.6 | Enough | TBD | DNA is much more degraded than the December fragments |
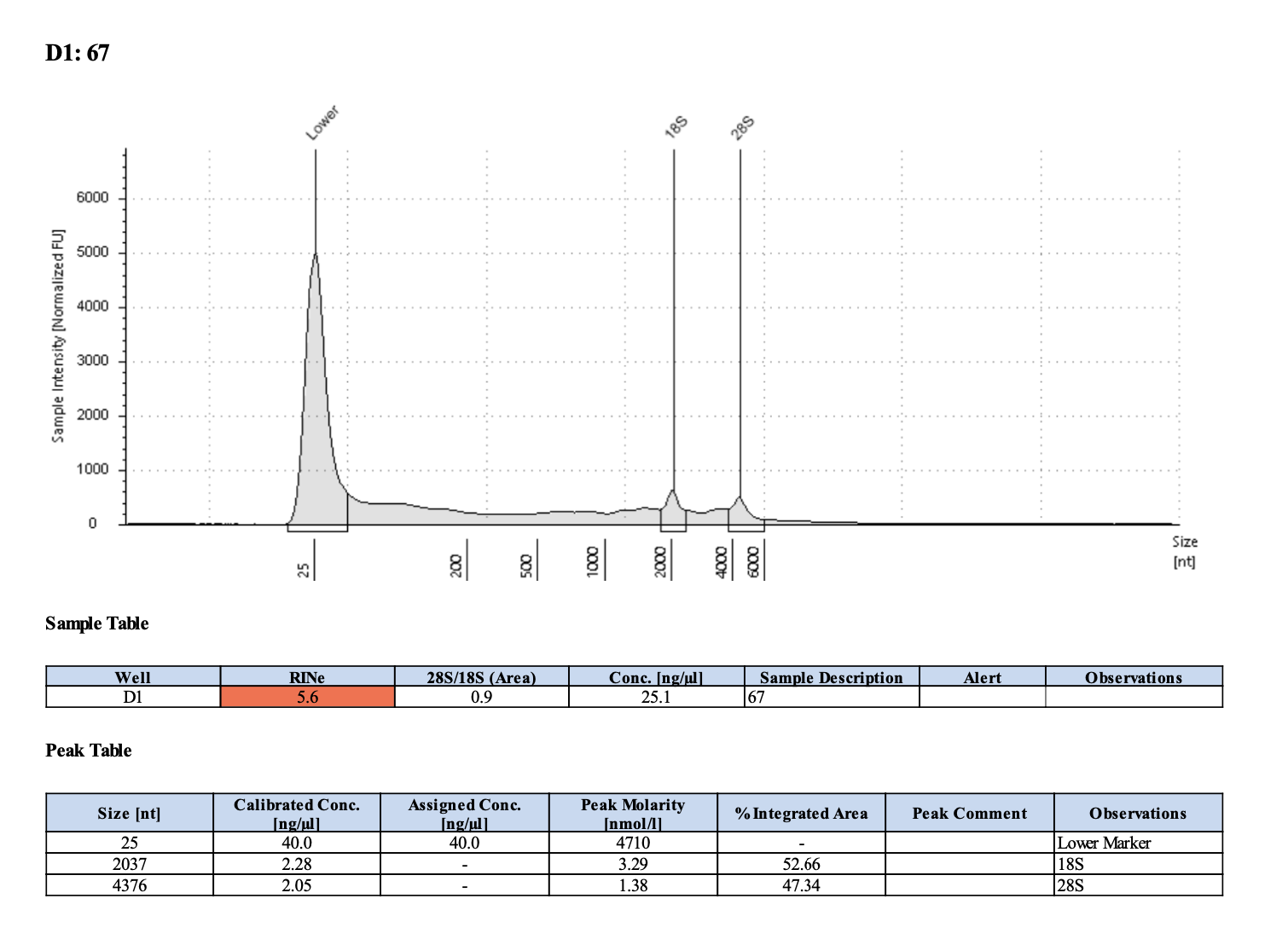
| M-3 | 2019-07-20 | 67 | 20211029 | 62.8 | 62.6 | 62.7 | 27 | 27 | 27 | 8.9 | Enough | Yes | DNA is much more degraded than the December fragments |
| M-20 | 2019-07-20 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | Couldn’t find this fragment - we may not have any left. Just send the RNA we have from the first round |
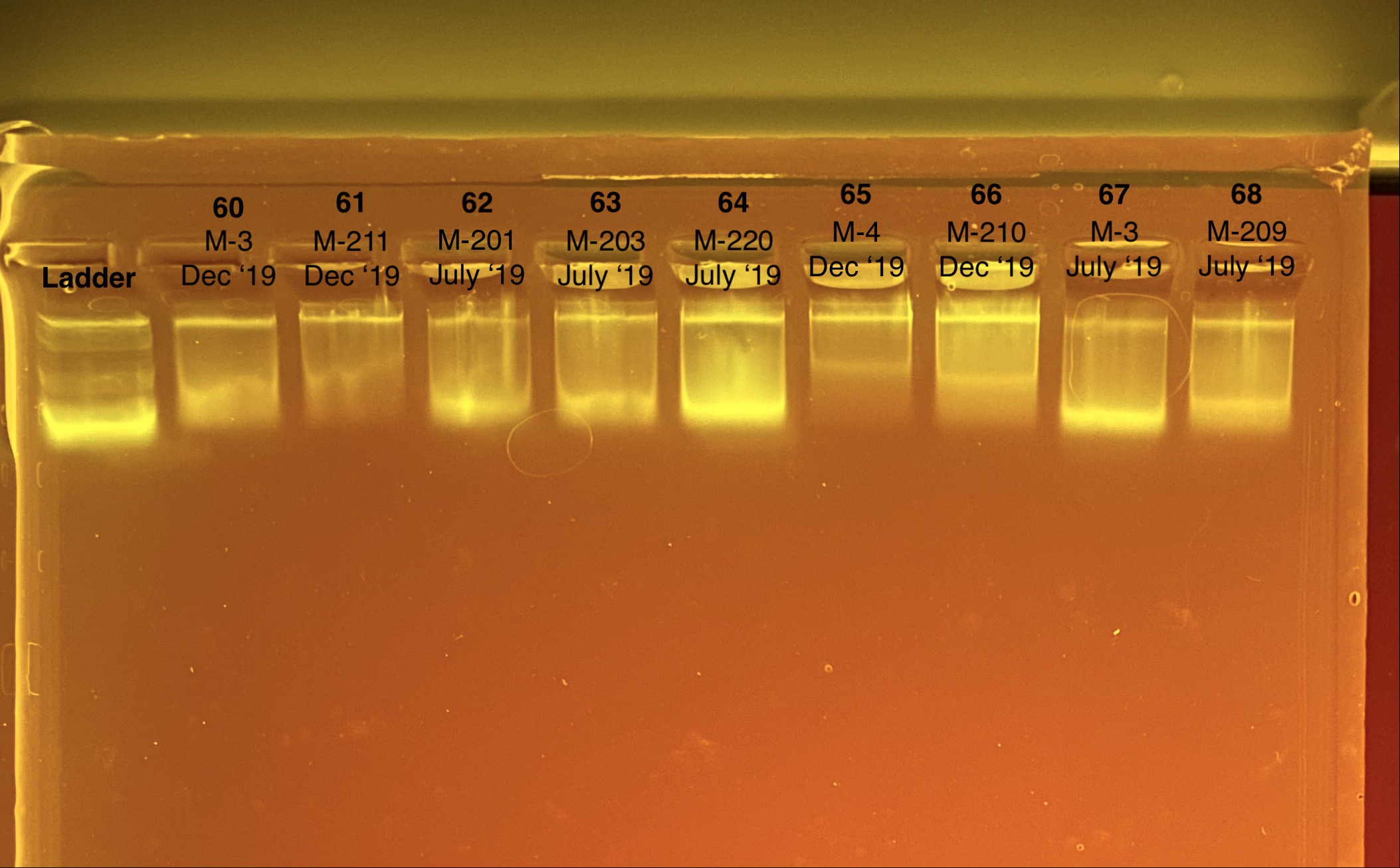
Tapestation links: Ext 60-64, Ext 65-68