mtORF Amplification Pocillopora
mtORF Amplification for Pocillopora spp. corals
Amplification of the mitochondria open reading frame (mtORF) region from DNA extracted from Pocillopora coral fragments from the Holobiont Integration 2018 project.
Following the Putnam Lab’s mtORF Amplification and Sanger Sequencing Prep Protocol.
Master spreadsheet for Holobiont Integration mtORF samples here.
10 ng/uL Dilutions
Each sample is diluted to 10 ng/uL concentration to standardize how much DNA we start with prior to PCR reaction.
| Plug_ID | Extraction Date | Species | Hard DNA (ng_ul) | DNA for dilution (ul) | Water for dilution (ul) | Notes |
|---|---|---|---|---|---|---|
| 1041 | 20190823 | Pocillopora | 44 | 2.27 | 7.73 | ✓ |
| 1043 | 20190814 | Pocillopora | 25.9 | 3.86 | 6.14 | ✓ |
| 1047 | 20190808 | Pocillopora | 37.4 | 2.67 | 7.33 | ✓ |
| 1050 | 20190809 | Pocillopora | 35.4 | 2.82 | 7.18 | ✓ |
| 1051 | 20190731 | Pocillopora | 45.5 | 2.20 | 7.80 | ✓ |
| 1059 | 20190826 | Pocillopora | 25.2 | 3.97 | 6.03 | ✓ |
| 1060 | 20190725 | Pocillopora | 44.3 | 2.26 | 7.74 | ✓ |
| 1090 | 20190807 | Pocillopora | 14 | 7.14 | 2.86 | ✓ |
| 1103 | 20190718 | Pocillopora | 50.9 | 1.96 | 8.04 | ✓ |
| 1131 | 20190718 | Pocillopora | 30.5 | 3.28 | 6.72 | ✓ |
| 1138 | 20190809 | Pocillopora | 33.1 | 3.02 | 6.98 | ✓ |
| 1141 | 20190722 | Pocillopora | 24.1 | 4.15 | 5.85 | ✓ |
| 1147 | 20191206 | Pocillopora | 39.3 | 2.54 | 7.46 | ✓ |
| 1159 | 20190720 | Pocillopora | 4.38 | 22.83 | -12.83 | ✓ |
| 1168 | 20190730 | Pocillopora | 21.5 | 4.65 | 5.35 | ✓ |
| 1184 | 20190731 | Pocillopora | 33.7 | 2.97 | 7.03 | ✓ |
| 1205 | 20190720 | Pocillopora | 27.3 | 3.66 | 6.34 | ✓ |
| 1207 | 20190826 | Pocillopora | 35.9 | 2.79 | 7.21 | ✓ |
| 1219 | 20190724 | Pocillopora | 40.2 | 2.49 | 7.51 | ✓ |
| 1220 | 20190730 | Pocillopora | 20.3 | 4.93 | 5.07 | ✓ |
| 1225 | 20190730 | Pocillopora | 21.6 | 4.63 | 5.37 | ✓ |
| 1227 | 20190801 | Pocillopora | 10.55 | 9.48 | 0.52 | ✓ |
| 1238 | 20190725 | Pocillopora | 27.4 | 3.65 | 6.35 | ✓ |
| 1239 | 20190823 | Pocillopora | 28.6 | 3.50 | 6.50 | ✓ |
| 1254 | 20190731 | Pocillopora | 32.7 | 3.06 | 6.94 | ✓ |
| 1281 | 20190924 | Pocillopora | 16.7 | 5.99 | 4.01 | ✓ |
| 1296 | 20190729 | Pocillopora | 21.5 | 4.65 | 5.35 | ✓ |
| 1302 | 20191111 | Pocillopora | 15.55 | 6.43 | 3.57 | ✓ |
| 1303 | 20190726 | Pocillopora | 18.85 | 5.31 | 4.69 | ✓ |
| 1329 | 20191204 | Pocillopora | 48.1 | 2.08 | 7.92 | ✓ |
| 1330 | 20190801 | Pocillopora | 16.8 | 5.95 | 4.05 | ✓ |
| 1343 | 20190814 | Pocillopora | 11.7 | 8.55 | 1.45 | ✓ |
| 1415 | 20190815 | Pocillopora | 36.4 | 2.75 | 7.25 | ✓ |
| 1416 | 20191009 | Pocillopora | 7.1 | 14.08 | -4.08 | ✓ |
| 1418 | 20190720 | Pocillopora | 31.2 | 3.21 | 6.79 | ✓ |
| 1427 | 20190807 | Pocillopora | 29.7 | 3.37 | 6.63 | ✓ |
| 1445 | 20190826 | Pocillopora | 26.1 | 3.83 | 6.17 | ✓ |
| 1451 | 20190720 | Pocillopora | 39.5 | 2.53 | 7.47 | ✓ |
| 1459 | 20190722 | Pocillopora | 50 | 2.00 | 8.00 | ✓ |
| 1466 | 20190731 | Pocillopora | 32.8 | 3.05 | 6.95 | ✓ |
| 1468 | 20191121 | Pocillopora | 15.95 | 6.27 | 3.73 | ✓ |
| 1471 | 20190718 | Pocillopora | 41.2 | 2.43 | 7.57 | ✓ |
| 1486 | 20190720 | Pocillopora | 37.2 | 2.69 | 7.31 | ✓ |
| 1487 | 20190807 | Pocillopora | 36.1 | 2.77 | 7.23 | ✓ |
| 1536 | 20190725 | Pocillopora | 58.2 | 1.72 | 8.28 | ✓ |
| 1542 | 20190815 | Pocillopora | 15.35 | 6.51 | 3.49 | ✓ |
| 1559 | 20190725 | Pocillopora | 35.4 | 2.82 | 7.18 | ✓ |
| 1563 | 20190905 | Pocillopora | 24 | 4.17 | 5.83 | ✓ |
| 1571 | 20191121 | Pocillopora | 25.7 | 3.89 | 6.11 | ✓ |
| 1581 | 20190826 | Pocillopora | 37.2 | 2.69 | 7.31 | ✓ |
| 1582 | 20190926 | Pocillopora | 20.9 | 4.78 | 5.22 | ✓ |
| 1594 | 20190720 | Pocillopora | 16.1 | 6.21 | 3.79 | ✓ |
| 1595 | 20190808 | Pocillopora | 57.6 | 1.74 | 8.26 | ✓ |
| 1596 | 20191009 | Pocillopora | 13.1 | 7.63 | 2.37 | ✓ |
| 1617 | 20190801 | Pocillopora | 29.9 | 3.34 | 6.66 | ✓ |
| 1637 | 20190805 | Pocillopora | 49.6 | 2.02 | 7.98 | ✓ |
| 1641 | 20190720 | Pocillopora | 26.9 | 3.72 | 6.28 | ✓ |
| 1642 | 20190903 | Pocillopora | 37.3 | 2.68 | 7.32 | ✓ |
| 1647 | 20190926 | Pocillopora | 59.3 | 1.69 | 8.31 | ✓ |
| 1653 | 20190826 | Pocillopora | 31.9 | 3.13 | 6.87 | ✓ |
| 1676 | 20190718 | Pocillopora | 23.5 | 4.26 | 5.74 | ✓ |
| 1696 | 20190718 | Pocillopora | 23.9 | 4.18 | 5.82 | ✓ |
| 1701 | 20190826 | Pocillopora | 34.2 | 2.92 | 7.08 | ✓ |
| 1707 | 20190905 | Pocillopora | 35.4 | 2.82 | 7.18 | ✓ |
| 1709 | 20191121 | Pocillopora | 20 | 5.00 | 5.00 | ✓ |
| 1721 | 20190805 | Pocillopora | 19.35 | 5.17 | 4.83 | ✓ |
| 1728 | 20190806 | Pocillopora | 48.1 | 2.08 | 7.92 | ✓ |
| 1732 | 20190724 | Pocillopora | 42.6 | 2.35 | 7.65 | ✓ |
| 1744 | 20190807 | Pocillopora | 21.8 | 4.59 | 5.41 | ✓ |
| 1755 | 20190801 | Pocillopora | 12 | 8.33 | 1.67 | ✓ |
| 1757 | 20190731 | Pocillopora | 31.4 | 3.18 | 6.82 | ✓ |
| 1762 | 20191121 | Pocillopora | 18.85 | 5.31 | 4.69 | ✓ |
| 1765 | 20190725 | Pocillopora | 50.5 | 1.98 | 8.02 | ✓ |
| 1767 | 20190823 | Pocillopora | 52.1 | 1.92 | 8.08 | ✓ |
| 1775 | 20190718 | Pocillopora | 37.2 | 2.69 | 7.31 | ✓ |
| 1777 | 20190930 | Pocillopora | 39.2 | 2.55 | 7.45 | ✓ |
| 1820 | 20190815 | Pocillopora | 21.4 | 4.67 | 5.33 | ✓ |
| 2002 | 20190826 | Pocillopora | 26.5 | 3.77 | 6.23 | ✓ |
| 2005 | 20190718 | Pocillopora | 30.3 | 3.30 | 6.70 | ✓ |
| 2012 | 20190730 | Pocillopora | 36.1 | 2.77 | 7.23 | ✓ |
| 2026 | 20190718 | Pocillopora | 14.85 | 6.73 | 3.27 | ✓ |
| 2064 | 20190730 | Pocillopora | 29.5 | 3.39 | 6.61 | ✓ |
| 2072 | 20190814 | Pocillopora | 20.2 | 4.95 | 5.05 | ✓ |
| 2087 | 20190826 | Pocillopora | 35.3 | 2.83 | 7.17 | ✓ |
| 2185 | 20191009 | Pocillopora | 15.9 | 6.29 | 3.71 | ✓ |
| 2195 | 20191121 | Pocillopora | 30 | 3.33 | 6.67 | ✓ |
| 2197 | 20190729 | Pocillopora | 25.4 | 3.94 | 6.06 | ✓ |
| 2202 | 20190724 | Pocillopora | 50.8 | 1.97 | 8.03 | ✓ |
| 2210 | 20190826 | Pocillopora | 21.4 | 4.67 | 5.33 | ✓ |
| 2212 | 20190903 | Pocillopora | 19.3 | 5.18 | 4.82 | ✓ |
| 2300 | 20190731 | Pocillopora | 33.3 | 3.00 | 7.00 | ✓ |
| 2304 | 20190722 | Pocillopora | 12.75 | 7.84 | 2.16 | ✓ |
| 2305 | 20190724 | Pocillopora | 83 | 1.20 | 8.80 | ✓ |
| 2306 | 20190926 | Pocillopora | 53.8 | 1.86 | 8.14 | ✓ |
| 2357 | 20190718 | Pocillopora | 61.8 | 1.62 | 8.38 | ✓ |
| 2363 | 20190720 | Pocillopora | 40.2 | 2.49 | 7.51 | ✓ |
| 2409 | 20190808 | Pocillopora | 55.7 | 1.80 | 8.20 | ✓ |
| 2413 | 20190809 | Pocillopora | 36.9 | 2.71 | 7.29 | ✓ |
| 2414 | 20190807 | Pocillopora | 48 | 2.08 | 7.92 | ✓ |
| 2513 | 20190722 | Pocillopora | 12.55 | 7.97 | 2.03 | ✓ |
| 2527 | 20190805 | Pocillopora | 61.2 | 1.63 | 8.37 | ✓ |
| 2534 | 20191208 | Pocillopora | 48.7 | 2.05 | 7.95 | ✓ |
| 2550 | 20190724 | Pocillopora | 60.4 | 1.66 | 8.34 | ✓ |
| 2564 | 20190726 | Pocillopora | 57.5 | 1.74 | 8.26 | ✓ |
| 2668 | 20191001 | Pocillopora | 31.1 | 3.22 | 6.78 | ✓ |
| 2733 | 20191113 | Pocillopora | 32.8 | 3.05 | 6.95 | ✓ |
| 2743 | 20190724 | Pocillopora | 101 | 0.99 | 9.01 | ✓ |
| 2750 | 20190905 | Pocillopora | 45.8 | 2.18 | 7.82 | ✓ |
| 2861 | 20190730 | Pocillopora | 25.3 | 3.95 | 6.05 | ✓ |
| 2870 | 20190801 | Pocillopora | 36 | 2.78 | 7.22 | ✓ |
| 2873 | 20190730 | Pocillopora | 44.2 | 2.26 | 7.74 | ✓ |
| 2877 | 20190731 | Pocillopora | 59.1 | 1.69 | 8.31 | ✓ |
| 2878 | 20190807 | Pocillopora | 48.7 | 2.05 | 7.95 | ✓ |
| 2879 | 20190930 | Pocillopora | 33.5 | 2.99 | 7.01 | ✓ |
| 2977 | 20190815 | Pocillopora | 14.75 | 6.78 | 3.22 | ✓ |
| 2979 | 20190722 | Pocillopora | 32.2 | 3.11 | 6.89 | ✓ |
| 2993 | 20190725 | Pocillopora | 48.4 | 2.07 | 7.93 | ✓ |
| 2999 | 20190801 | Pocillopora | 44 | 2.27 | 7.73 | ✓ |
| 1169 | 20191113 | Pocillopora | 28.4 | 3.52 | 6.48 | ✓ |
Notes:
- 1755: 7.6 uL of DNA and 2.4 uL of water added instead of original value; double check at qubit step that this isn’t too diluted. In the end we only need a small amount of DNA so it might be okay. Otherwise I can re-do this sample at the end.
- All dilutions done on 20200914
- In gel look for 1459 to make sure it has DNA, the 2.00 uL DNA addition was checked off (probably just missed physically checking this off on my printed sheet); same for 1775 and 2409
- 1169 done on 20201102-3 because it was originally left off the list. All other dilutions made at one time.
Dilution Plates
PLATE 1:
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 1254 | 1676 | 2979 | 1471 | NA | 1757 | 2861 | 1238 | 1536 | 1418 | 1141 | 1219 |
| B | 1103 | 2363 | 2304 | 1594 | 2999 | 2357 | 1765 | NA | 1184 | 2993 | 1205 | 2513 |
| C | 2870 | 1227 | 1159 | 1296 | 1451 | 2026 | 1732 | 1644 | 2202 | 1459 | 2300 | 2305 |
| D | 1168 | 2005 | 2873 | 1134 | 2064 | 1051 | 1617 | 2564 | 2550 | 1330 | 2877 | 1721 |
| E | 1486 | 1303 | 1696 | 1225 | 1775 | 1466 | NA | 1220 | 2012 | 1755 | 2195 | 1302 |
| F | 1468 | 2733 | 1329 | 1239 | 1762 | 1709 | 1571 | 1147 | 2534 | 1207 | 2527 | 2668 |
| G | 2087 | 1653 | 2414 | 1415 | 1820 | 1637 | 1445 | 2002 | 1138 | 1595 | 1041 | 1090 |
| H | 1744 | 1343 | 1767 | 2878 | 2977 | 1043 | 1582 | 1281 | 1701 | 2210 | NA | 1581 |
PLATE 2:
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 1647 | 1563 | 1542 | 1059 | 2306 | 1487 | 1047 | 2212 | 2409 | 1050 | 2413 | 1707 |
| B | 2879 | 1777 | 2185 | 1427 | 2072 | 1728 | 2750 | 1416 | 1642 | 1596 | 1060 | 2197 |
| C | 2743 | 1559 | 1169 | |||||||||
| D | ||||||||||||
| E | ||||||||||||
| F | ||||||||||||
| G | ||||||||||||
| H |
Notes:
- 1581 has higher volume of liquid when I looked at the dilution plate. Check in qubit step that is sample isn’t too diluted.
- Plates are labeled on the side and with a foil seal on top. Stored at -20C.
PCR Reactions
PCR Plate Outline
Samples are put in columns 1, 4, 7, and 10 so that triplicates can be made in the adjacent two columns using the multi-chanel pipette (ex: 1254 will be in B1, B2, and B3 during the PCR run). See the protocol for details on running in triplicates.
Plate 1 - PCR made and run on 20200915
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | Neg Control | 1536 | 2999 | 1227 | ||||||||
| B | 1254 | 1418 | 2357 | 1159 | ||||||||
| C | 1676 | 1141 | 1765 | 1296 | ||||||||
| D | 2979 | 1219 | 1184 | 1451 | ||||||||
| E | 1471 | 1103 | 2993 | 2026 | ||||||||
| F | 1757 | 2363 | 1205 | 1732 | ||||||||
| G | 2861 | 2304 | 2513 | 1641 | ||||||||
| H | 1238 | 1594 | 2870 | Neg Control |
Plate 2 - PCR made and run on 20200915
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | Neg Control | 2064 | 1486 | 1755 | ||||||||
| B | 1459 | 1051 | 1303 | 2195 | ||||||||
| C | 2300 | 1617 | 1696 | 1302 | ||||||||
| D | 2305 | 2564 | 1225 | 1468 | ||||||||
| E | 1168 | 2550 | 1775 | 2733 | ||||||||
| F | 2005 | 1330 | 1466 | 1329 | ||||||||
| G | 2873 | 2877 | 1220 | 1239 | ||||||||
| H | 1131 | 1721 | 2012 | Neg control |
Plate 3
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | Neg Control | 2087 | 1138 | 2977 | ||||||||
| B | 1709 | 1653 | 1595 | 1043 | ||||||||
| C | 1571 | 2414 | 1041 | 1582 | ||||||||
| D | 1147 | 1415 | 1090 | 1281 | ||||||||
| E | 2534 | 1820 | 1744 | 1701 | ||||||||
| F | 1207 | 1637 | 1343 | 2210 | ||||||||
| G | 2527 | 1445 | 1767 | 1581 | ||||||||
| H | 2668 | 2002 | 2878 | Neg control |
Plate 4
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | Neg Control | 2409 | 2072 | 2743 | ||||||||
| B | 1563 | 1050 | 1728 | 1559 | ||||||||
| C | 1542 | 2413 | 2750 | 2202 | ||||||||
| D | 1059 | 1707 | 1416 | 1762 | ||||||||
| E | 2306 | 2879 | 1642 | 1647 | ||||||||
| F | 1487 | 1777 | 1596 | Neg control | ||||||||
| G | 1047 | 2185 | 1060 | Neg control | 1169 | |||||||
| H | 2212 | 1427 | 2197 | Neg control | 1755 |
Sanity check: 30 samples for Plate 1, 2, and 3 (90 total), 28 samples for Plate 4 = 118 total samples.
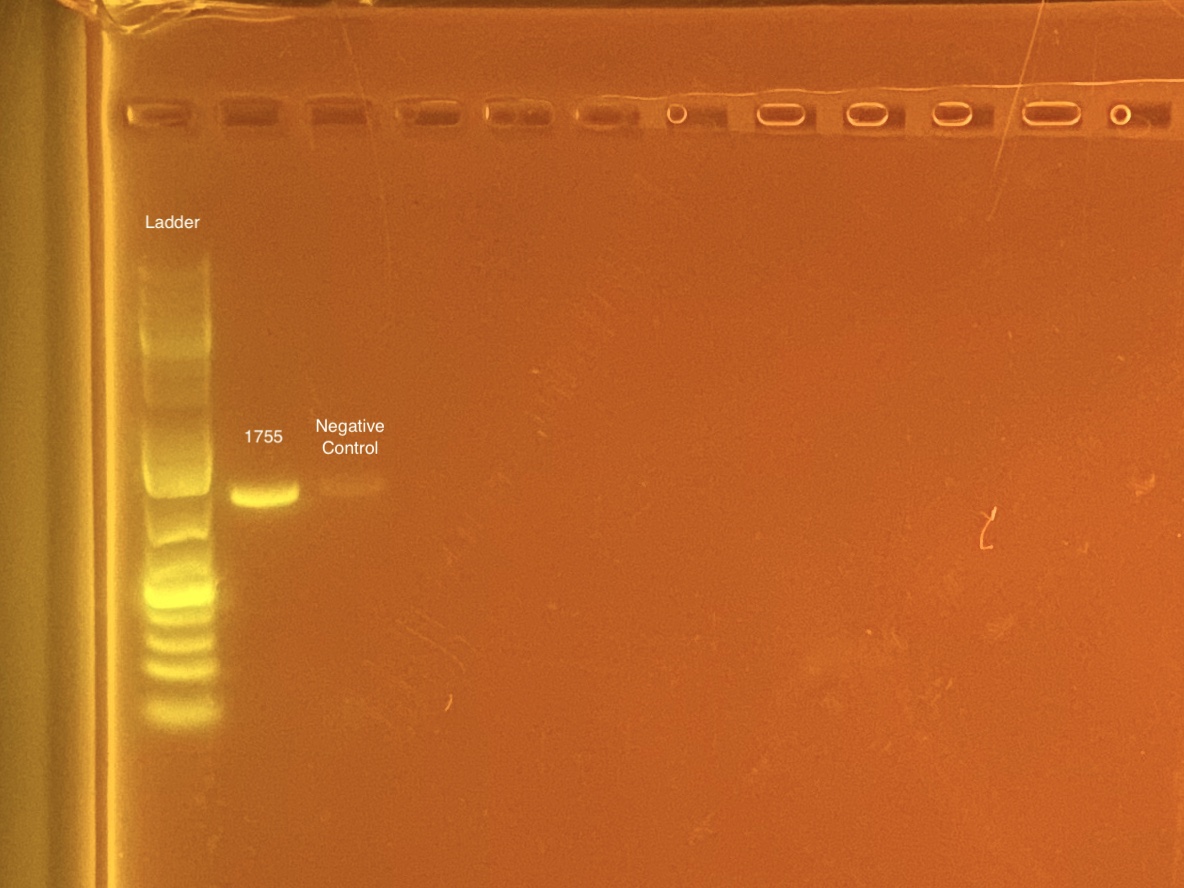
STRIP TUBE 1 - 20201028 1755; Neg control
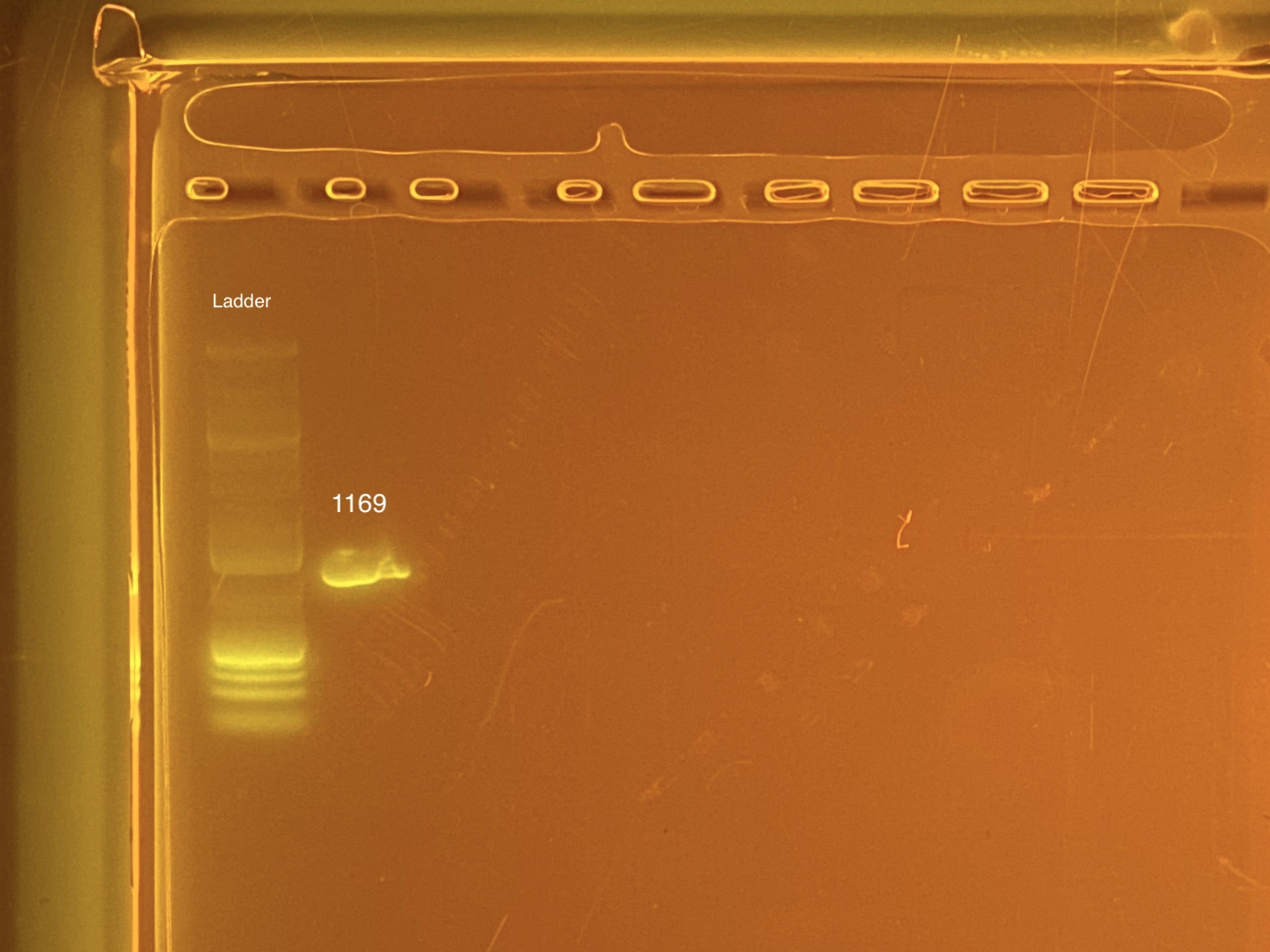
STRIP TUBE 2 - 20201102 1169; Neg control
Notes:
- If I have a full dilution plate (like plate 1), I should have had the PCR plate be identical to the columns in the full dilution plate so that I can multi-chanel pipette
- 1755 re-done at later date. The 1755 that worked is in PCR plate 4 well H12.
- 1169 was done at a later date because it was originally left off of the list. Well G12.
PCR Runs 20200915 - Plates 1 and 2 run
Master mix created for half of the samples. 60 samples, 4 negative controls (blanks), and 1 for potential pipetting error.
| Master Mix | uL | # of samples (60 + 4 neg controls + 5 for error) | total needed (uL) |
|---|---|---|---|
| Phusion PCR master mix | 50 | 69 | 3450 |
| UltraPure water | 44 | 69 | 3036 |
| 10uM working stock FatP6.1 primer | 1.3 | 69 | 89.70 |
| 10uM working stock RORF primer | 1.3 | 69 | 89.70 |
Sanity check that calculations are correct.
total volume MM = 6665.4
/ 97 uL per well
= 68.71546392 (number should equal calculated sample number above)
20201020 - Plates 3 and 4 run
Master mix created for half of the samples. 60 samples, 4 negative controls (blanks), and 1 for potential pipetting error.
| Master Mix | uL | # of samples (60 + 4 neg controls + 5 for error) | total needed (uL) |
|---|---|---|---|
| Phusion PCR master mix | 50 | 69 | 3450 |
| UltraPure water | 44 | 69 | 3036 |
| 10uM working stock FatP6.1 primer | 1.3 | 69 | 89.70 |
| 10uM working stock RORF primer | 1.3 | 69 | 89.70 |
Sanity check that calculations are correct.
total volume MM = 6665.4
/ 97 uL per well
= 68.71546392 (number should equal calculated sample number above)
20201028 - 1 sample and 1 control in a strip tube
| Master Mix | uL | # of samples (1 + 1 control + 0.1 error) | total needed (uL) |
|---|---|---|---|
| Phusion PCR master mix | 50 | 2.1 | 105 |
| UltraPure water | 44 | 2.1 | 92.4 |
| 10uM working stock FatP6.1 primer | 1.3 | 2.1 | 2.73 |
| 10uM working stock RORF primer | 1.3 | 2.1 | 2.73 |
Sanity check that calculations are correct.
total volume MM = 202.86
/ 97 uL per well = 2.09 (should be equal to sample number above)
20201102 - 1 sample and 1 control in a strip tube
| Master Mix | uL | # of samples (1 + 1 control + 0.1 error) | total needed (uL) |
|---|---|---|---|
| Phusion PCR master mix | 50 | 2.1 | 105 |
| UltraPure water | 44 | 2.1 | 92.4 |
| 10uM working stock FatP6.1 primer | 1.3 | 2.1 | 2.73 |
| 10uM working stock RORF primer | 1.3 | 2.1 | 2.73 |
Sanity check that calculations are correct.
total volume MM = 202.86
/ 97 uL per well = 2.09 (should be equal to sample number above)
1X Bead Clean-Up
20200915 Plates 1 and 2 cleaned. Stored in -20C freezer waiting to quantified.
No notes necessary, protocol followed exactly (see link at the top of this post).
End result is 50 uL of sample in each well that look identical to the PCR platemaps above.
20201025 Plates 3 and 4 cleaned. No notes; see above.
20201028: 1755 bead clean-up: No notes. 20201103 1169 bead clean up: No notes.
All stored in -20C freezer when not in use.
Qubit (ng/uL) Values
20200922: BR DNA Standard 1: 172.65 Standard 2: 16,875.12.
- Original standard 2 was 12,416.92 but re-made because thought that was much lower than normal.
20201026: BR DNA Standard 1: 200.79 Standard 2: 22,255.60.
20201028: BR DNA Standard 1: 184.39 Standard 2: 20,275.06.
20201103: BR DNA Standard 1: 218.44 Standard 2: 24,795.11.
| Qubit Date | Sample ID | DNA 1 | DNA 2 | Average DNA | Gel Date | Gel Pass? |
|---|---|---|---|---|---|---|
| 20200922 | 1594 | 6.7 | 6.76 | 6.73 | 2020923 | Yes |
| 20200922 | 1205 | 37.2 | 36.8 | 37 | 2020923 | Yes |
| 20200922 | 2513 | 26.4 | 26.2 | 26.3 | 2020923 | Yes |
| 20200922 | 2005 | 11.9 | 11.8 | 11.85 | 2020923 | Yes |
| 20200922 | 1732 | 20 | 19.9 | 19.95 | 2020923 | Yes |
| 20200922 | 1721 | 15.2 | 15.1 | 15.15 | 2020923 | Yes |
| 20200922 | 2733 | 19.7 | 19.5 | 19.6 | 2020923 | Yes |
| 20200922 | 1254 | 12.9 | 12.8 | 12.85 | 20201026 | Yes |
| 20200922 | 1676 | 19.4 | 19.2 | 19.3 | 20201026 | Yes |
| 20201026 | 2979 | 18.1 | 17.9 | 18 | 20201026 | Yes |
| 20200922 | 1471 | 19.2 | 19 | 19.1 | 20201026 | Yes |
| 20200922 | 1757 | 17.3 | 17.2 | 17.25 | 20201026 | Yes |
| 20200922 | 2861 | 20.2 | 20 | 20.1 | 20201026 | Yes |
| 20200922 | 1238 | 19.8 | 19.7 | 19.75 | 20201026 | Yes |
| 20200922 | 1536 | 9.46 | 9.32 | 9.39 | 20201026 | Yes |
| 20200922 | 1418 | 8.8 | 8.74 | 8.77 | 20201026 | Yes |
| 20200922 | 1141 | 8.74 | 8.66 | 8.7 | 20201026 | Yes |
| 20200922 | 1219 | 10.6 | 10.6 | 10.6 | 20201026 | Yes |
| 20200922 | 1103 | 8.46 | 8.38 | 8.42 | 20201026 | Yes |
| 20200922 | 2304 | 5.3 | 5.22 | 5.26 | 20201026 | Yes |
| 20200922 | 2999 | 23.8 | 23.6 | 23.7 | 20201026 | Yes |
| 20200922 | 2357 | 24.8 | 24.6 | 24.7 | 20201026 | Yes |
| 20200922 | 1765 | 26 | 25.8 | 25.9 | 20201026 | Yes |
| 20200922 | 1184 | 29.8 | 29.6 | 29.7 | 20201026 | Yes |
| 20200922 | 2993 | 34 | 33.6 | 33.8 | 20201026 | Yes |
| 20200922 | 2366 | 8.8 | 8.52 | 8.66 | 20201026 | Yes |
| 20200922 | 1159 | 31.8 | 31.4 | 31.6 | 20201026 | Yes |
| 20200922 | 1296 | 31.2 | 31 | 31.1 | 20201026 | Yes |
| 20200922 | 1451 | 34.8 | 34.4 | 34.6 | 20201026 | Yes |
| 20200922 | 1227 | 32 | 31.6 | 31.8 | 20201026 | Yes |
| 20200922 | 2026 | 32.2 | 31.8 | 32 | 20201026 | Yes |
| 20200922 | 2870 | 24.6 | 24.4 | 24.5 | 20201026 | Yes |
| 20200922 | 1459 | 14.8 | 14.6 | 14.7 | 20201026 | Yes |
| 20200922 | 2873 | 16.9 | 16.5 | 16.7 | 20201026 | Yes |
| 20200922 | 2300 | 20 | 19.8 | 19.9 | 20201026 | Yes |
| 20200922 | 1641 | 27.4 | 27.2 | 27.3 | 20201026 | Yes |
| 20200922 | 2305 | 15.1 | 15 | 15.05 | 20201026 | Yes |
| 20200922 | 1168 | 15.7 | 15.5 | 15.6 | 20201026 | Yes |
| 20200922 | 1131 | 17.2 | 17 | 17.1 | 20201026 | Yes |
| 20200922 | 2877 | 19.7 | 19.5 | 19.6 | 20201026 | Yes |
| 20200922 | 1617 | 30 | 29.8 | 29.9 | 20201026 | Yes |
| 20200922 | 2550 | 28.6 | 28.4 | 28.5 | 20201026 | Yes |
| 20200922 | 2064 | 32.4 | 32.2 | 32.3 | 20201026 | Yes |
| 20200922 | 1330 | 13.7 | 13.6 | 13.65 | 20201026 | Yes |
| 20200922 | 1051 | 26.6 | 26.4 | 26.5 | 20201026 | Yes |
| 20200922 | 2564 | 22.4 | 21.8 | 22.1 | 20201026 | Yes |
| 20200922 | 1696 | 14.5 | 14.2 | 14.35 | 20201026 | Yes |
| 20201026 | 1225 | 16.6 | 16.1 | 16.35 | 20201026 | Yes |
| 20200922 | 1486 | 20.8 | 20.6 | 20.7 | 20201026 | Yes |
| 20200922 | 1220 | 3.48 | 3.42 | 3.45 | 20201026 | Yes |
| 20200922 | 1466 | 12.2 | 12 | 12.1 | 20201026 | Yes |
| 20200922 | 1303 | 14.1 | 13.8 | 13.95 | 20201026 | Yes |
| 20200922 | 1775 | 15.2 | 15.1 | 15.15 | 20201026 | Yes |
| 20200922 | 1302 | 27.2 | 27 | 27.1 | 20201026 | Yes |
| 20200922 | 1329 | 24.2 | 24 | 24.1 | 20201026 | Yes |
| 20200922 | 1468 | 26 | 25.8 | 25.9 | 20201026 | Yes |
| 20201028 | 1755 | 7.2 | 7.14 | 7.17 | 20201028 | Yes |
| 20200922 | 1239 | 23.2 | 23 | 23.1 | 20201026 | Yes |
| 20200922 | 2012 | 22.8 | 22.6 | 22.7 | 20201026 | Yes |
| 20200922 | 2195 | 20.8 | 20.6 | 20.7 | 20201026 | Yes |
| 20201026 | 2534 | 15.9 | 15.4 | 15.65 | 20201026 | Yes |
| 20201026 | 1709 | 13.5 | 13.1 | 13.3 | 20201026 | Yes |
| 20201026 | 1571 | 12.1 | 11.7 | 11.9 | 20201026 | Yes |
| 20201026 | 2527 | 17.5 | 16.8 | 17.15 | 20201026 | Yes |
| 20201026 | 1147 | 14.9 | 14.5 | 14.7 | 20201026 | Yes |
| 20201026 | 2668 | 17.5 | 16.9 | 17.2 | 20201026 | Yes |
| 20201026 | 1207 | 17 | 16.4 | 16.7 | 20201026 | Yes |
| 20201026 | 1445 | 7.26 | 6.96 | 7.11 | 20201026 | Yes |
| 20201026 | 2414 | 6.86 | 6.58 | 6.72 | 20201026 | Yes |
| 20201026 | 2002 | 7.7 | 7.36 | 7.53 | 20201026 | Yes |
| 20201026 | 1820 | 4.84 | 4.66 | 4.75 | 20201026 | Yes |
| 20201026 | 1637 | 6.44 | 6.08 | 6.26 | 20201026 | Yes |
| 20201026 | 2087 | 5.68 | 5.48 | 5.58 | 20201026 | Yes |
| 20201026 | 1415 | 5.4 | 5.12 | 5.26 | 20201026 | Yes |
| 20201026 | 1653 | 7.58 | 7.3 | 7.44 | 20201026 | Yes |
| 20201026 | 1041 | 10.2 | 9.84 | 10.02 | 20201026 | Yes |
| 20201026 | 1595 | 12.8 | 12.6 | 12.7 | 20201026 | Yes |
| 20201026 | 1138 | 6.08 | 5.94 | 6.01 | 20201026 | Yes |
| 20201026 | 1090 | 11.6 | 11.3 | 11.45 | 20201026 | Yes |
| 20201026 | 1744 | 13.5 | 13.3 | 13.4 | 20201026 | Yes |
| 20201026 | 1343 | 8.92 | 8.68 | 8.8 | 20201026 | Yes |
| 20201026 | 1767 | 14.6 | 14.2 | 14.4 | 20201026 | Yes |
| 20201026 | 2878 | 15.1 | 14.8 | 14.95 | 20201026 | Yes |
| 20201026 | 1582 | 19.9 | 19.5 | 19.7 | 20201026 | Yes |
| 20201026 | 1045 | 19.9 | 19.1 | 19.5 | 20201026 | Yes |
| 20201026 | 2977 | 17.9 | 17.4 | 17.65 | 20201026 | Yes |
| 20201026 | 1281 | 23.4 | 22.8 | 23.1 | 20201026 | Yes |
| 20201026 | 1701 | 26.2 | 25.8 | 26 | 20201026 | Yes |
| 20201026 | 2210 | 23.2 | 22.6 | 22.9 | 20201026 | Yes |
| 20201026 | 1581 | 24.6 | 24.2 | 24.4 | 20201026 | Yes |
| 20201026 | 1060 | 19.2 | 18.4 | 18.8 | 20201026 | Yes |
| 20201026 | 2197 | 18 | 17.4 | 17.7 | 20201026 | Yes |
| 20201026 | 2202 | 8.64 | 8.22 | 8.43 | 20201026 | Yes |
| 20201026 | 2743 | 8.92 | 8.6 | 8.76 | 20201026 | Yes |
| 20201026 | 1762 | 10.9 | 10.6 | 10.75 | 20201026 | Yes |
| 20201026 | 1559 | 8.84 | 8.38 | 8.61 | 20201026 | Yes |
| 20201026 | 1487 | 19.7 | 19 | 19.35 | 20201026 | Yes |
| 20201026 | 1047 | 22.4 | 21.4 | 21.9 | 20201026 | Yes |
| 20201026 | 2409 | 12.7 | 12.2 | 12.45 | 20201026 | Yes |
| 20201026 | 1647 | 10.5 | 10.3 | 10.4 | 20201026 | Yes |
| 20201026 | 2212 | 19.7 | 19 | 19.35 | 20201026 | Yes |
| 20201026 | 2750 | 16.2 | 15.7 | 15.95 | 20201026 | Yes |
| 20201026 | 2185 | 12.2 | 11.1 | 11.65 | 20201026 | Yes |
| 20201026 | 1059 | 22.8 | 22 | 22.4 | 20201026 | Yes |
| 20201026 | 2072 | 15.7 | 15.3 | 15.5 | 20201026 | Yes |
| 20201026 | 1563 | 19.4 | 18.5 | 18.95 | 20201026 | Yes |
| 20201026 | 1642 | 15.6 | 15.1 | 15.35 | 20201026 | Yes |
| 20201026 | 1596 | 19 | 18.4 | 18.7 | 20201026 | Yes |
| 20201026 | 1050 | 13.2 | 12.9 | 13.05 | 20201026 | Yes |
| 20201026 | 1728 | 16.2 | 15.3 | 15.75 | 20201026 | Yes |
| 20201026 | 1777 | 12.5 | 12.2 | 12.35 | 20201026 | Yes |
| 20201026 | 1427 | 13.9 | 13.5 | 13.7 | 20201026 | Yes |
| 20201026 | 1416 | 16.3 | 15.7 | 16 | 20201026 | Yes |
| 20201026 | 2879 | 13.3 | 13 | 13.15 | 20201026 | Yes |
| 20201026 | 1707 | 13.6 | 13.3 | 13.45 | 20201026 | Yes |
| 20201103 | 1169 | 15.6 | 15.5 | 15.55 | 20201103 | Yes |
Those with ** for values were too low to measure. I will re-qubit these in case 1 full uL wasn’t added to the qubit tube. Otherwise there might not have been a full uL added to the PCR plate.
Gel Electrophoresis Analysis
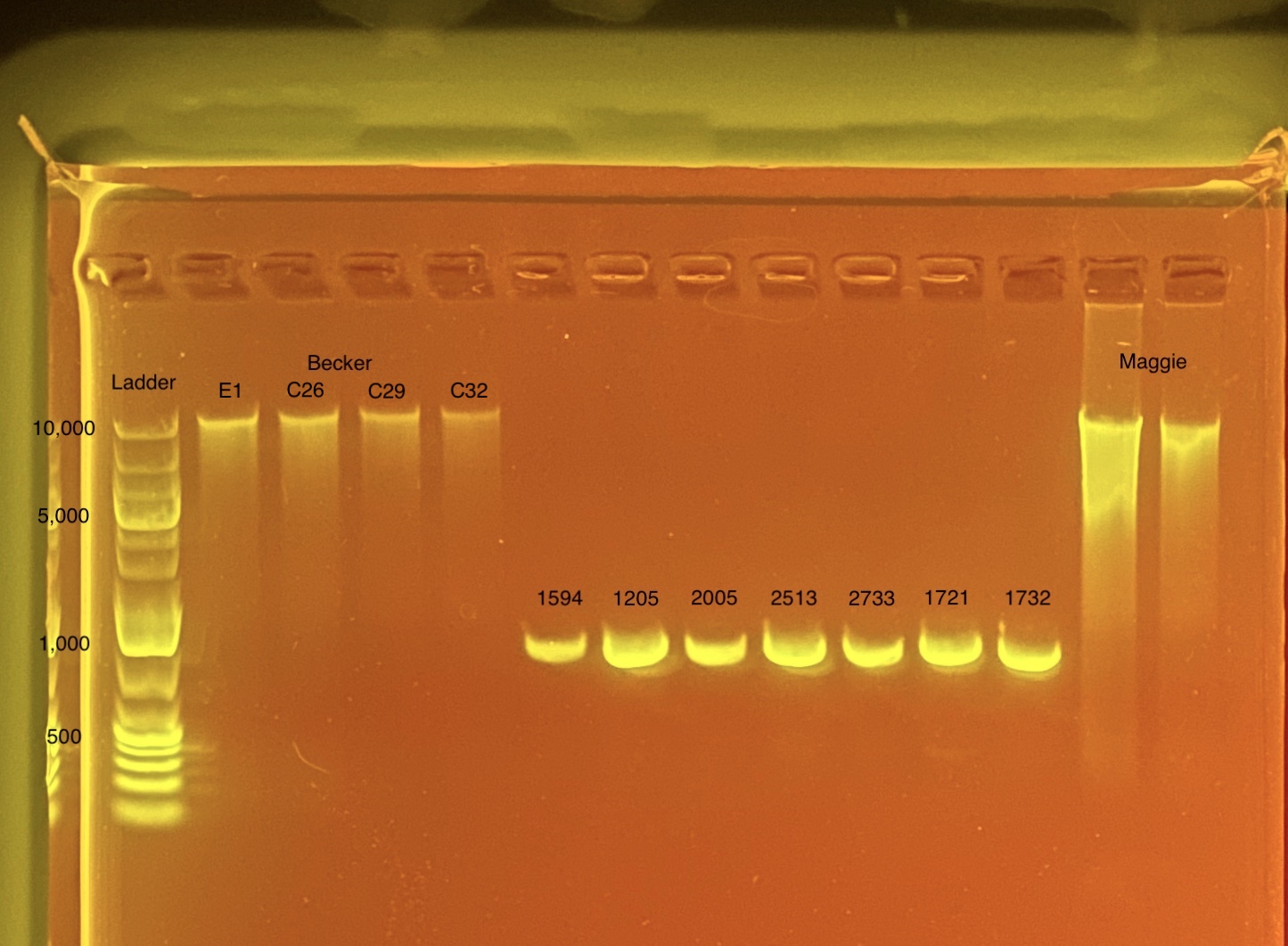
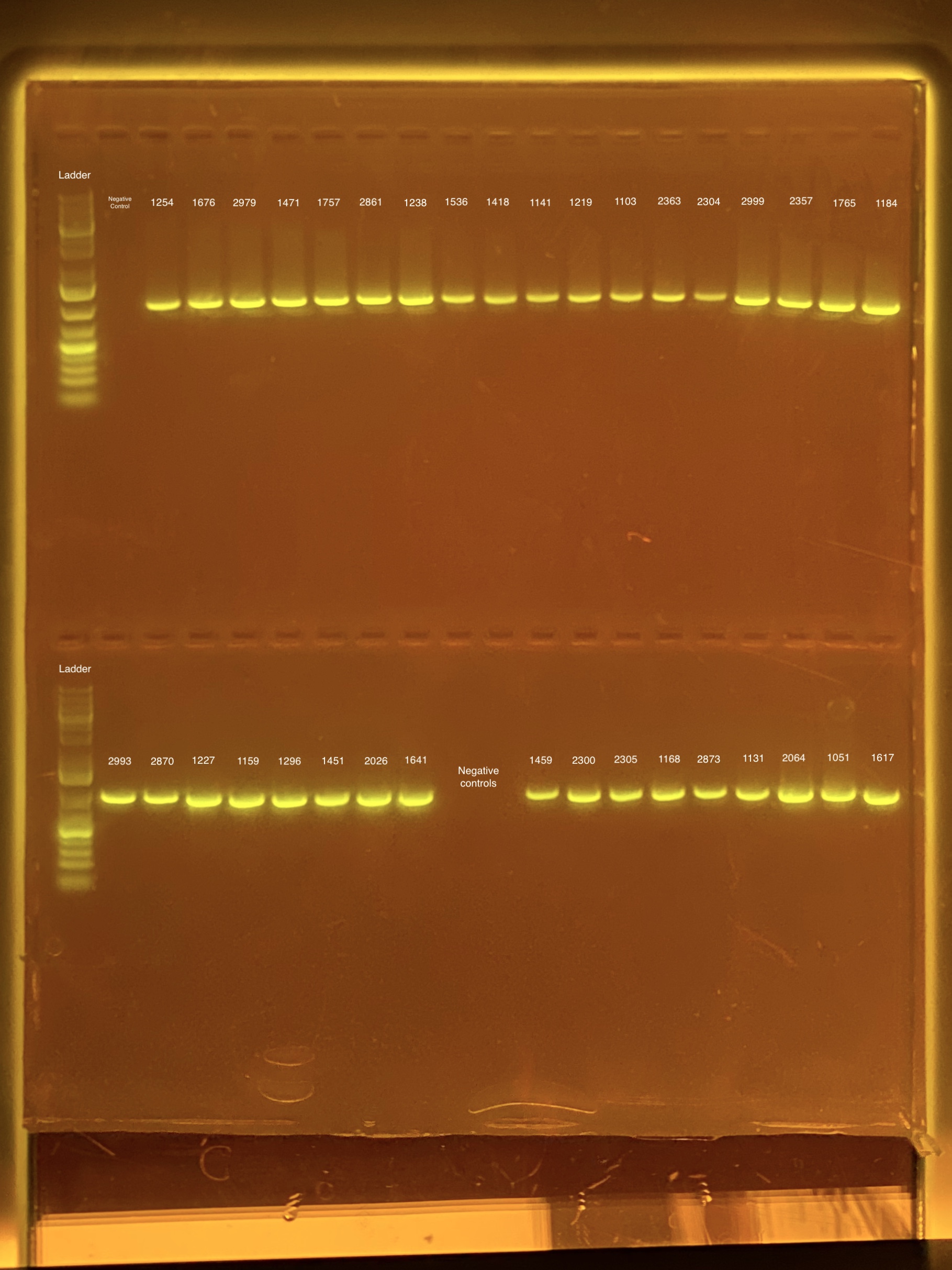
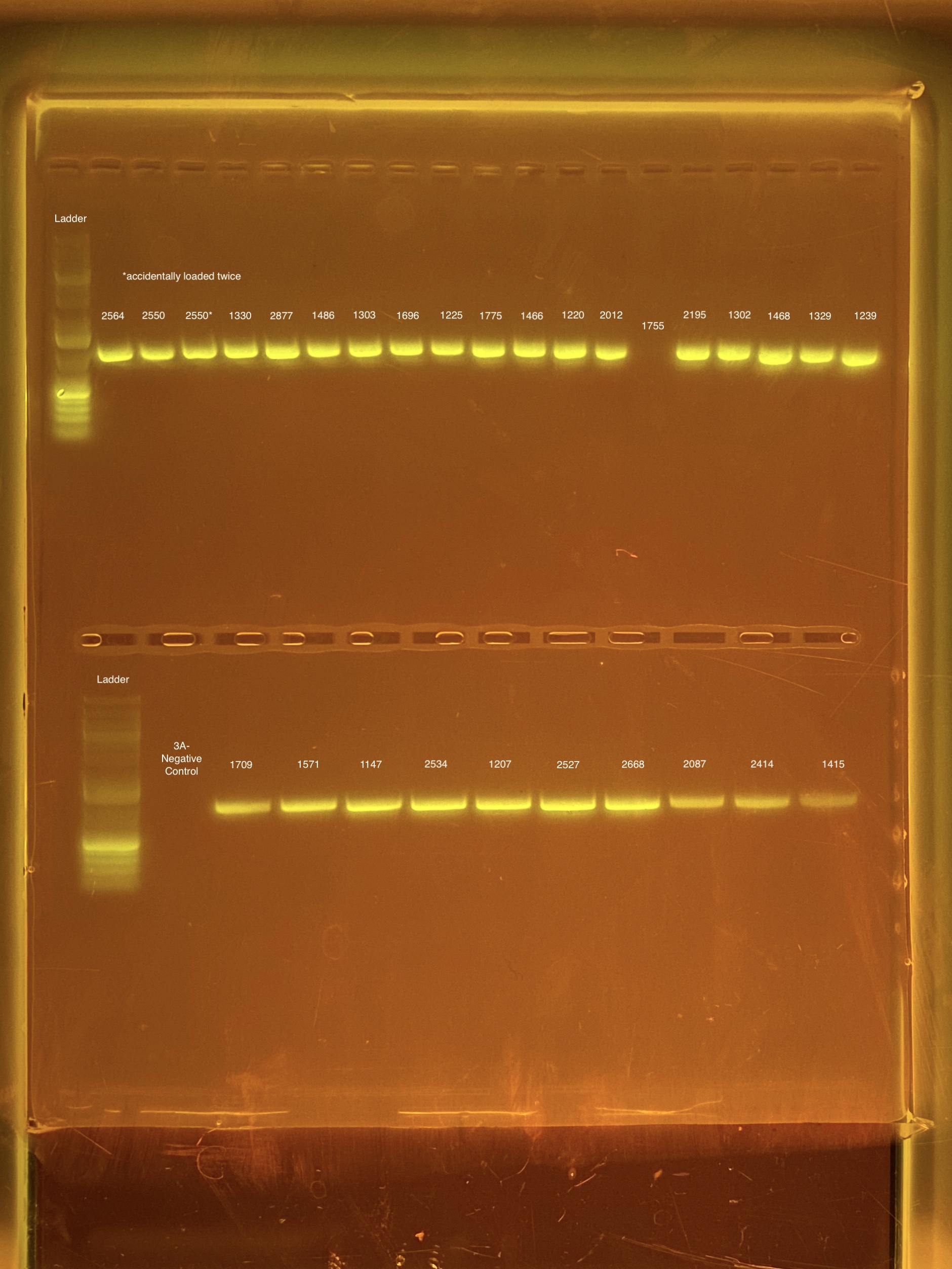
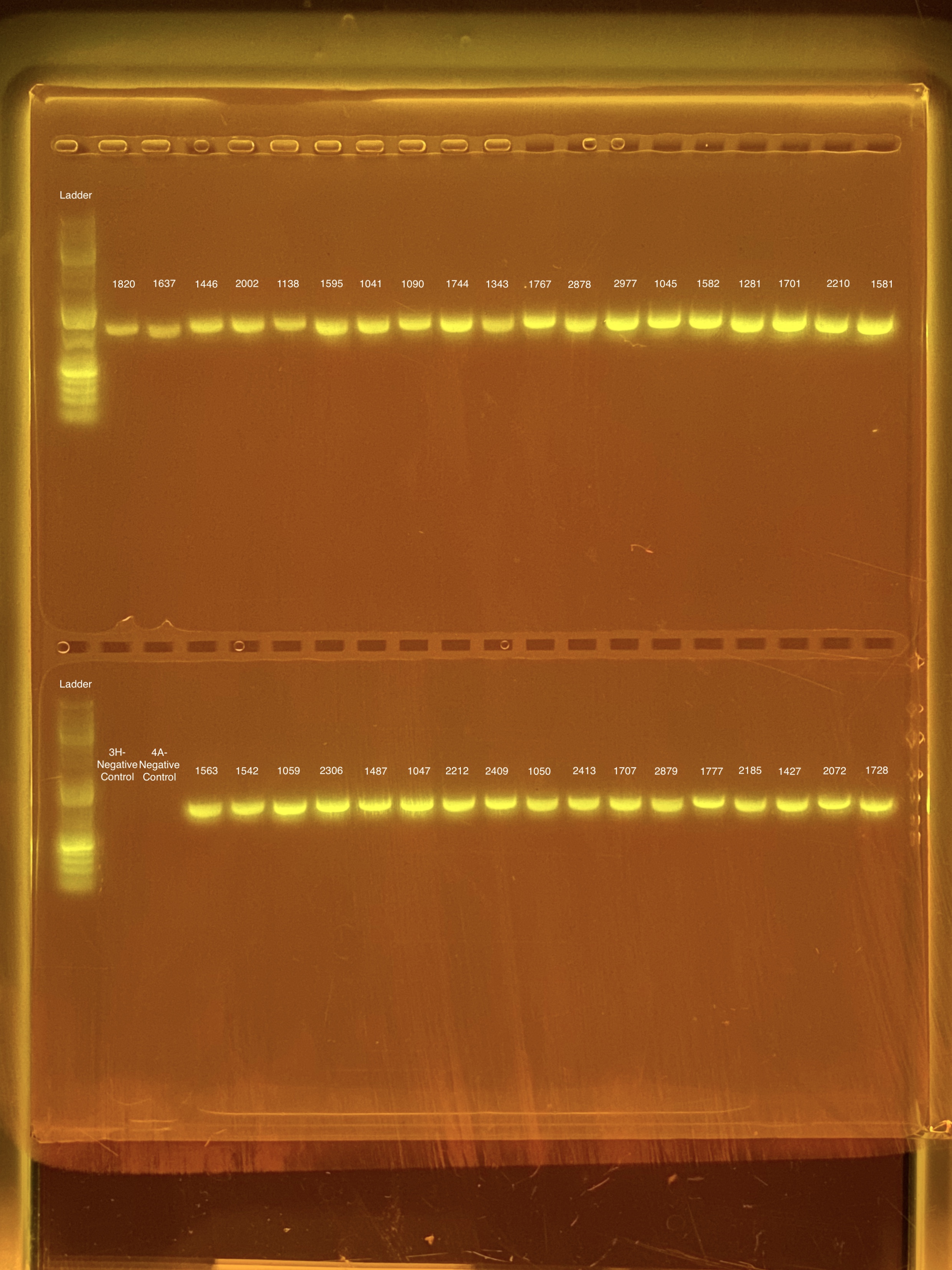
mtORF amplication will result in a sharp band ~1,000 bp.
20200923 Gel:
20201026 Gels:
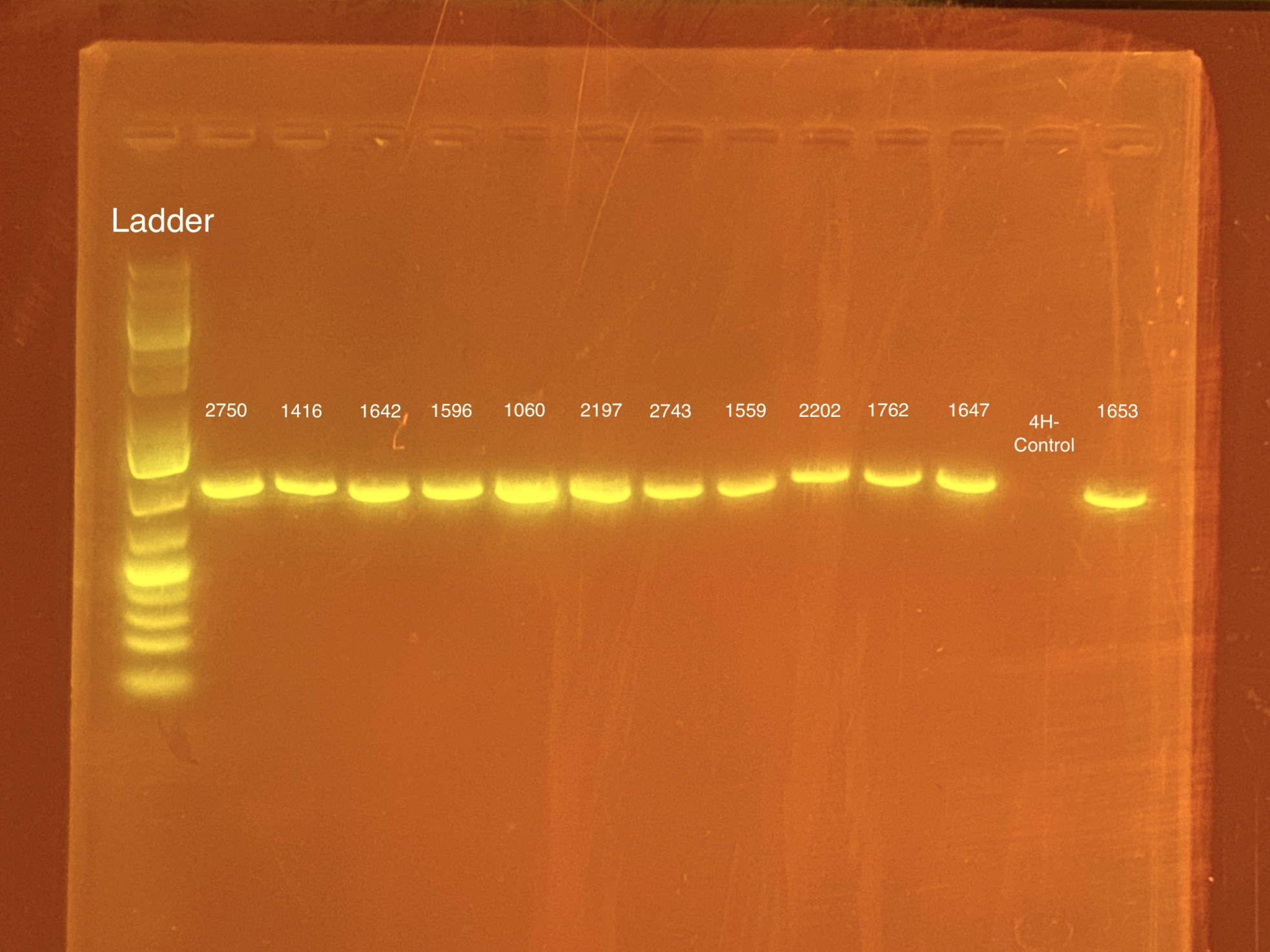
20201028 Gel:
20201103 Gel:
Sequencing Prep
20200923: A small set of 7 samples (HPE01-07) were chosen to analyze on a gel before sending to sequence as a test run. These samples were chosen randomly but I tried to make sure a range of qubit values were included (6 - 30 ng) and samples from both PCR plates were included from various columns.
20201103: The rest of the samples were dropped off for sequencing (HPE08-119).
1:2 dilution chosen; 5 uL of DNA and 5 uL of water added.
| Prep Date | Sample ID | DNA 1 | DNA 2 | Average DNA(ng_uL) | 1:2 Dilutions (ng_uL) | DNA to 25 ng | H2O to 10 uL | Notes |
|---|---|---|---|---|---|---|---|---|
| 20200924 | 1205 | 37.2 | 36.8 | 37 | 18.50 | 1.35 | 8.65 | ✓ |
| 20200924 | 1594 | 6.7 | 6.76 | 6.73 | 3.37 | 7.43 | 2.57 | ✓ |
| 20200924 | 1721 | 15.2 | 15.1 | 15.15 | 7.58 | 3.30 | 6.70 | ✓ |
| 20200924 | 1732 | 20 | 19.9 | 19.95 | 9.98 | 2.51 | 7.49 | ✓ |
| 20200924 | 2005 | 11.9 | 11.8 | 11.85 | 5.93 | 4.22 | 5.78 | ✓ |
| 20200924 | 2513 | 26.4 | 26.2 | 26.3 | 13.15 | 1.90 | 8.10 | ✓ |
| 20200924 | 2733 | 19.7 | 19.5 | 19.6 | 9.80 | 2.55 | 7.45 | ✓ |
| 20201102 | 1169 | 15.5 | 15.6 | 15.55 | 7.78 | 3.22 | 6.78 | ✓ |
| 20201028-9 | 1041 | 10.2 | 9.84 | 10.02 | 5.01 | 4.99 | 5.01 | ✓ |
| 20201028-9 | 1043 | 26 | 26 | 26 | 13.00 | 1.92 | 8.08 | ✓ |
| 20201028-9 | 1047 | 22.4 | 21.4 | 21.9 | 10.95 | 2.28 | 7.72 | ✓ |
| 20201028-9 | 1050 | 13.2 | 12.9 | 13.05 | 6.53 | 3.83 | 6.17 | ✓ |
| 20201028-9 | 1051 | 26.6 | 26.4 | 26.5 | 13.25 | 1.89 | 8.11 | ✓ |
| 20201028-9 | 1059 | 22.8 | 22 | 22.4 | 11.20 | 2.23 | 7.77 | ✓ |
| 20201028-9 | 1060 | 19.2 | 18.4 | 18.8 | 9.40 | 2.66 | 7.34 | ✓ |
| 20201028-9 | 1090 | 11.6 | 11.3 | 11.45 | 5.73 | 4.37 | 5.63 | ✓ |
| 20201028-9 | 1103 | 8.46 | 8.38 | 8.42 | 4.21 | 5.94 | 4.06 | ✓ |
| 20201028-9 | 1131 | 17.2 | 17 | 17.1 | 8.55 | 2.92 | 7.08 | ✓ |
| 20201028-9 | 1138 | 6.08 | 5.94 | 6.01 | 3.01 | 8.32 | 1.68 | ✓ |
| 20201028-9 | 1141 | 8.74 | 8.66 | 8.7 | 4.35 | 5.75 | 4.25 | ✓ |
| 20201028-9 | 1147 | 14.9 | 14.5 | 14.7 | 7.35 | 3.40 | 6.60 | ✓ |
| 20201028-9 | 1159 | 31.8 | 31.4 | 31.6 | 15.80 | 1.58 | 8.42 | ✓ |
| 20201028-9 | 1168 | 15.7 | 15.5 | 15.6 | 7.80 | 3.21 | 6.79 | ✓ |
| 20201028-9 | 1184 | 29.8 | 29.6 | 29.7 | 14.85 | 1.68 | 8.32 | ✓ |
| 20201028-9 | 1207 | 17 | 16.4 | 16.7 | 8.35 | 2.99 | 7.01 | ✓ |
| 20201028-9 | 1219 | 10.6 | 10.6 | 10.6 | 5.30 | 4.72 | 5.28 | ✓ |
| 20201028-9 | 1220 | 3.48 | 3.42 | 3.45 | 3.435 | 7.25 | 2.75 | Not diluted; ✓ |
| 20201028-9 | 1225 | 16.6 | 16.1 | 16.35 | 8.18 | 3.06 | 6.94 | ✓ |
| 20201028-9 | 1227 | 32 | 31.6 | 31.8 | 15.90 | 1.57 | 8.43 | ✓ |
| 20201028-9 | 1238 | 19.8 | 19.7 | 19.75 | 9.88 | 2.53 | 7.47 | ✓ |
| 20201028-9 | 1239 | 23.2 | 23 | 23.1 | 11.55 | 2.16 | 7.84 | ✓ |
| 20201028-9 | 1254 | 12.9 | 12.8 | 12.85 | 6.43 | 3.89 | 6.11 | ✓ |
| 20201028-9 | 1281 | 23.4 | 22.8 | 23.1 | 11.55 | 2.16 | 7.84 | ✓ |
| 20201028-9 | 1296 | 31.2 | 31 | 31.1 | 15.55 | 1.61 | 8.39 | ✓ |
| 20201028-9 | 1302 | 27.2 | 27 | 27.1 | 13.55 | 1.85 | 8.15 | ✓ |
| 20201028-9 | 1303 | 14.1 | 13.8 | 13.95 | 6.98 | 3.58 | 6.42 | ✓ |
| 20201028-9 | 1329 | 24.2 | 24 | 24.1 | 12.05 | 2.07 | 7.93 | ✓ |
| 20201028-9 | 1330 | 13.7 | 13.6 | 13.65 | 6.83 | 3.66 | 6.34 | ✓ |
| 20201028-9 | 1343 | 8.92 | 8.68 | 8.8 | 4.40 | 5.68 | 4.32 | ✓ |
| 20201028-9 | 1415 | 5.4 | 5.12 | 5.26 | 2.63 | 9.51 | 0.49 | ✓ |
| 20201028-9 | 1416 | 16.3 | 15.7 | 16 | 8.00 | 3.13 | 6.88 | ✓ |
| 20201028-9 | 1418 | 8.8 | 8.74 | 8.77 | 4.39 | 5.70 | 4.30 | ✓ |
| 20201028-9 | 1427 | 13.9 | 13.5 | 13.7 | 6.85 | 3.65 | 6.35 | ✓ |
| 20201028-9 | 1445 | 7.26 | 6.96 | 7.11 | 3.56 | 7.03 | 2.97 | ✓ |
| 20201028-9 | 1451 | 34.8 | 34.4 | 34.6 | 17.30 | 1.45 | 8.55 | ✓ |
| 20201028-9 | 1459 | 14.8 | 14.6 | 14.7 | 7.35 | 3.40 | 6.60 | ✓ |
| 20201028-9 | 1466 | 12.2 | 12 | 12.1 | 6.05 | 4.13 | 5.87 | ✓ |
| 20201028-9 | 1468 | 26 | 25.8 | 25.9 | 12.95 | 1.93 | 8.07 | ✓ |
| 20201028-9 | 1471 | 19.2 | 19 | 19.1 | 9.55 | 2.62 | 7.38 | ✓ |
| 20201028-9 | 1486 | 20.8 | 20.6 | 20.7 | 10.35 | 2.42 | 7.58 | ✓ |
| 20201028-9 | 1487 | 19.7 | 19 | 19.35 | 9.68 | 2.58 | 7.42 | ✓ |
| 20201028-9 | 1536 | 9.46 | 9.32 | 9.39 | 4.70 | 5.32 | 4.68 | ✓ |
| 20201028-9 | 1542 | 19.4 | 19.4 | 19.4 | 9.70 | 2.58 | 7.42 | ✓ |
| 20201028-9 | 1559 | 8.84 | 8.38 | 8.61 | 4.31 | 5.81 | 4.19 | ✓ |
| 20201028-9 | 1563 | 19.4 | 18.5 | 18.95 | 9.48 | 2.64 | 7.36 | ✓ |
| 20201028-9 | 1571 | 12.1 | 11.7 | 11.9 | 5.95 | 4.20 | 5.80 | ✓ |
| 20201028-9 | 1581 | 24.6 | 24.2 | 24.4 | 12.20 | 2.05 | 7.95 | ✓ |
| 20201028-9 | 1582 | 19.9 | 19.5 | 19.7 | 9.85 | 2.54 | 7.46 | ✓ |
| 20201028-9 | 1595 | 12.8 | 12.6 | 12.7 | 6.35 | 3.94 | 6.06 | ✓ |
| 20201028-9 | 1596 | 19 | 18.4 | 18.7 | 9.35 | 2.67 | 7.33 | ✓ |
| 20201028-9 | 1617 | 30 | 29.8 | 29.9 | 14.95 | 1.67 | 8.33 | ✓ |
| 20201028-9 | 1637 | 6.44 | 6.08 | 6.26 | 3.13 | 7.99 | 2.01 | ✓ |
| 20201028-9 | 1641 | 27.4 | 27.2 | 27.3 | 13.65 | 1.83 | 8.17 | ✓ |
| 20201028-9 | 1642 | 15.6 | 15.1 | 15.35 | 7.68 | 3.26 | 6.74 | ✓ |
| 20201028-9 | 1647 | 10.5 | 10.3 | 10.4 | 5.20 | 4.81 | 5.19 | ✓ |
| 20201028-9 | 1653 | 7.58 | 7.3 | 7.44 | 3.72 | 6.72 | 3.28 | ✓ |
| 20201028-9 | 1676 | 19.4 | 19.2 | 19.3 | 9.65 | 2.59 | 7.41 | ✓ |
| 20201028-9 | 1696 | 14.5 | 14.2 | 14.35 | 7.18 | 3.48 | 6.52 | ✓ |
| 20201028-9 | 1701 | 26.2 | 25.8 | 26 | 13.00 | 1.92 | 8.08 | ✓ |
| 20201028-9 | 1707 | 13.6 | 13.3 | 13.45 | 6.73 | 3.72 | 6.28 | ✓ |
| 20201028-9 | 1709 | 13.5 | 13.1 | 13.3 | 6.65 | 3.76 | 6.24 | ✓ |
| 20201028-9 | 1728 | 16.2 | 15.3 | 15.75 | 7.88 | 3.17 | 6.83 | ✓ |
| 20201028-9 | 1744 | 13.5 | 13.3 | 13.4 | 6.70 | 3.73 | 6.27 | ✓ |
| 20201028-9 | 1755 | 7.2 | 7.14 | 7.17 | 3.59 | 6.97 | 3.03 | ✓ |
| 20201028-9 | 1757 | 17.3 | 17.2 | 17.25 | 8.63 | 2.90 | 7.10 | ✓ |
| 20201028-9 | 1762 | 10.9 | 10.6 | 10.75 | 5.38 | 4.65 | 5.35 | ✓ |
| 20201028-9 | 1765 | 26 | 25.8 | 25.9 | 12.95 | 1.93 | 8.07 | ✓ |
| 20201028-9 | 1767 | 14.6 | 14.2 | 14.4 | 7.20 | 3.47 | 6.53 | ✓ |
| 20201028-9 | 1775 | 15.2 | 15.1 | 15.15 | 7.58 | 3.30 | 6.70 | ✓ |
| 20201028-9 | 1777 | 12.5 | 12.2 | 12.35 | 6.18 | 4.05 | 5.95 | ✓ |
| 20201028-9 | 1820 | 4.84 | 4.66 | 4.75 | 4.705 | 5.26 | 4.74 | Not diluted; ✓ |
| 20201028-9 | 2002 | 7.7 | 7.36 | 7.53 | 3.77 | 6.64 | 3.36 | ✓ |
| 20201028-9 | 2012 | 22.8 | 22.6 | 22.7 | 11.35 | 2.20 | 7.80 | ✓ |
| 20201028-9 | 2026 | 32.2 | 31.8 | 32 | 16.00 | 1.56 | 8.44 | ✓ |
| 20201028-9 | 2064 | 32.4 | 32.2 | 32.3 | 16.15 | 1.55 | 8.45 | ✓ |
| 20201028-9 | 2072 | 15.7 | 15.3 | 15.5 | 7.75 | 3.23 | 6.77 | ✓ |
| 20201028-9 | 2087 | 5.68 | 5.48 | 5.58 | 2.79 | 8.96 | 1.04 | ✓ |
| 20201028-9 | 2185 | 12.2 | 11.1 | 11.65 | 5.83 | 4.29 | 5.71 | ✓ |
| 20201028-9 | 2195 | 20.8 | 20.6 | 20.7 | 10.35 | 2.42 | 7.58 | ✓ |
| 20201028-9 | 2197 | 18 | 17.4 | 17.7 | 8.85 | 2.82 | 7.18 | ✓ |
| 20201028-9 | 2202 | 8.64 | 8.22 | 8.43 | 4.22 | 5.93 | 4.07 | ✓ |
| 20201028-9 | 2210 | 23.2 | 22.6 | 22.9 | 11.45 | 2.18 | 7.82 | ✓ |
| 20201028-9 | 2212 | 19.7 | 19 | 19.35 | 9.68 | 2.58 | 7.42 | ✓ |
| 20201028-9 | 2300 | 20 | 19.8 | 19.9 | 9.95 | 2.51 | 7.49 | ✓ |
| 20201028-9 | 2304 | 5.3 | 5.22 | 5.26 | 2.63 | 9.51 | 0.49 | ✓ |
| 20201028-9 | 2305 | 15.1 | 15 | 15.05 | 7.53 | 3.32 | 6.68 | ✓ |
| 20201028-9 | 2306 | 26.2 | 26 | 26.1 | 13.05 | 1.92 | 8.08 | ✓ |
| 20201028-9 | 2357 | 24.8 | 24.6 | 24.7 | 12.35 | 2.02 | 7.98 | ✓ |
| 20201028-9 | 2363 | 5.24 | 5.2 | 5.22 | 2.61 | 9.58 | 0.42 | ✓ |
| 20201028-9 | 2409 | 12.7 | 12.2 | 12.45 | 6.23 | 4.02 | 5.98 | ✓ |
| 20201028-9 | 2413 | 13.7 | 13.7 | 13.7 | 6.85 | 3.65 | 6.35 | ✓ |
| 20201028-9 | 2414 | 6.86 | 6.58 | 6.72 | 3.36 | 7.44 | 2.56 | ✓ |
| 20201028-9 | 2527 | 17.5 | 16.8 | 17.15 | 8.58 | 2.92 | 7.08 | ✓ |
| 20201028-9 | 2534 | 15.9 | 15.4 | 15.65 | 7.83 | 3.19 | 6.81 | ✓ |
| 20201028-9 | 2550 | 28.6 | 28.4 | 28.5 | 14.25 | 1.75 | 8.25 | ✓ |
| 20201028-9 | 2564 | 22.4 | 21.8 | 22.1 | 11.05 | 2.26 | 7.74 | ✓ |
| 20201028-9 | 2668 | 17.5 | 16.9 | 17.2 | 8.60 | 2.91 | 7.09 | ✓ |
| 20201028-9 | 2743 | 8.92 | 8.6 | 8.76 | 4.38 | 5.71 | 4.29 | ✓ |
| 20201028-9 | 2750 | 16.2 | 15.7 | 15.95 | 7.98 | 3.13 | 6.87 | ✓ |
| 20201028-9 | 2861 | 20.2 | 20 | 20.1 | 10.05 | 2.49 | 7.51 | ✓ |
| 20201028-9 | 2870 | 24.6 | 24.4 | 24.5 | 12.25 | 2.04 | 7.96 | ✓ |
| 20201028-9 | 2873 | 16.9 | 16.5 | 16.7 | 8.35 | 2.99 | 7.01 | ✓ |
| 20201028-9 | 2877 | 19.7 | 19.5 | 19.6 | 9.80 | 2.55 | 7.45 | ✓ |
| 20201028-9 | 2878 | 15.1 | 14.8 | 14.95 | 7.48 | 3.34 | 6.66 | ✓ |
| 20201028-9 | 2879 | 13.3 | 13 | 13.15 | 6.58 | 3.80 | 6.20 | ✓ |
| 20201028-9 | 2977 | 17.9 | 17.4 | 17.65 | 8.83 | 2.83 | 7.17 | ✓ |
| 20201028-9 | 2979 | 18.1 | 17.9 | 18 | 9.00 | 2.78 | 7.22 | ✓ |
| 20201028-9 | 2993 | 34 | 33.6 | 33.8 | 16.90 | 1.48 | 8.52 | ✓ |
| 20201028-9 | 2999 | 23.8 | 23.6 | 23.7 | 11.85 | 2.11 | 7.89 | ✓ |
| 20201028-9 | NA_1045 | 19.9 | 19.1 | 19.5 | 9.75 | 2.56 | 7.44 | Not on list? Qubit # miswritten |
| 20201028-9 | NA_2366 | 8.8 | 8.52 | 8.66 | 4.33 | 5.77 | 4.23 | Not on list? Qubit # miswritten |
See the Putnam lab mtORF protocol for the URI GSC sequencing forms.
1:2 Dilution platemaps
Plate 1
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | Neg Control | 1536 | 2999 | 1227 | Neg Control | 2064 | 1486 | 1755 | Neg Control | 2087 | 1138 | 2977 |
| B | 1254 | 1418 | 2357 | 1159 | 1459 | 1051 | 1303 | 2195 | 1709 | 1653 | 1595 | 1043 |
| C | 1676 | 1141 | 1765 | 1296 | 2300 | 1617 | 1696 | 1302 | 1571 | 2414 | 1041 | 1582 |
| D | 2979 | 1219 | 1184 | 1451 | 2305 | 2564 | 1225 | 1468 | 1147 | 1415 | 1090 | 1281 |
| E | 1471 | 1103 | 2993 | 2026 | 1168 | 2550 | 1775 | 2733 | 2534 | 1820 | 1744 | 1701 |
| F | 1757 | 2363 | 1205 | 1732 | 2005 | 1330 | 1466 | 1329 | 1207 | 1637 | 1343 | 2210 |
| G | 2861 | 2304 | 2513 | 1641 | 2873 | 2877 | 1220 | 1239 | 2527 | 1445 | 1767 | 1581 |
| H | 1238 | 1594 | 2870 | Neg Control | 1131 | 1721 | 2012 | Neg control | 2668 | 2002 | 2878 | Neg control |
Plate 2
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | Neg Control | 2409 | 2072 | 2743 | ||||||||
| B | 1563 | 1050 | 1728 | 1559 | ||||||||
| C | 1542 | 2413 | 2750 | 2202 | ||||||||
| D | 1059 | 1707 | 1416 | 1762 | ||||||||
| E | 2306 | 2879 | 1642 | 1647 | ||||||||
| F | 1487 | 1777 | 1596 | Neg control | ||||||||
| G | 1047 | 2185 | 1060 | 1755 | ||||||||
| H | 2212 | 1427 | 2197 | 1169 |
25 ng platemaps that were dropped off for sequencing
10 uL of 25ng DNA sample and 2 uL of 3.2 uM primer is in each well.
Plate 1
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 1254 | 1676 | 2179 | 1471 | 1757 | 2861 | 1238 | 1536 | 1418 | 1141 | 1219 | 1103 |
| B | 2304 | 2999 | 2357 | 1765 | 1184 | 2993 | 2870 | 1227 | 1159 | 1296 | 1451 | 2026 |
| C | 1641 | 1459 | 2300 | 2305 | 1168 | 2873 | 1131 | 2064 | 1051 | 1617 | 2564 | 2550 |
| D | 1330 | 2877 | 1486 | 1303 | 1696 | 1225 | 1775 | 1466 | 1220 | 2012 | 2195 | 1302 |
| E | 1468 | 1329 | 1239 | 1709 | 1571 | 1147 | 2534 | 1207 | 2527 | 2668 | 2087 | 1653 |
| F | 2414 | 1415 | 1820 | 1637 | 1445 | 2002 | 1138 | 1595 | 1041 | 1090 | 1744 | 1343 |
| G | 1767 | 2878 | 2977 | 1582 | 1281 | 1701 | 2210 | 1581 | 1563 | 1059 | 1487 | 1047 |
| H | 2212 | 2409 | 1050 | 1707 | 2879 | 1777 | 2185 | 1427 | 2072 | 1728 | 2750 | 1416 |
Plate 2
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 1642 | 2743 | ||||||||||
| B | 1542 | 1559 | ||||||||||
| C | 1596 | 2202 | ||||||||||
| D | 2306 | 1762 | ||||||||||
| E | 1060 | 1647 | ||||||||||
| F | 2413 | 1755 | ||||||||||
| G | 2197 | 2363 | ||||||||||
| H | 1169 | 1043 |
3.2 uM calculations
20201028:
3.2 uM primer = 68.48 uL 10 uM primer + 145.52 uL ultrapure water
20201103:
x = (3.2 uM Fat6.1 primer)(3 uL total)/10 uM Fat6.1 primer = 0.96 uL 10 uM Fat6.1 primer
3.2 uM primer = 0.96 uL 10 uM primer + 2.04 ultrapure water
2 samples did not run in sequencing
I uploaded sequences to the program Geneious to align all 119 sequences returned from GSC and determine which 2 didn’t work. HPE70 and HPE78 sequences were the two files with no sequences, which were 1581 and 1563.
My initial thought was that the primer wasn’t added correctly to the 25ng dilution plates that were given to GSC. So, I re-qubited the samples from the 1:2 dilution plates to make sure there was DNA added. Below are those results.
BR DNA Standard 1: 193.10
BR DNA Standard 2: 22,836.21
1581 DNA 1: 11.8, DNA 2: 11.7, Average: 11.75 ng/uL.
1563 DNA 1: 8.42, DNA 2: 8.32, Average: 8.37 ng/uL.
From this, it was most likely an issue with adding primer or DNA to the 25 ng plate.
25 ng plate calculations:
1581 (11.75 ng/uL): 2.13 uL DNA + 7.87 uL Ultrapure H2O + 2 uL 3.2 uM primer
1563 (8.32 ng/uL): 2.99 uL DNA + 7.01 uL Ultrapure H2O + 2 uL 3.2 uM primer
4 uL of 3.2 uM primer = 1.28 uL 10 uM primer + 2.72 uL Ultrapure H2O
HPE120 = 1581
HPE121 = 1563
These samples were re-made in PCR strip tubes labeled HPE120 and HPE121, then placed in Janet/GSC’s freezer on 20201111.
