WGBS Pico Methyl Seq Test Run
WGBS Pico Methylation Protocol Trials
Google spreadsheet with all WGBS raw data from E.Strand
As 20201112, two test runs have been done for WGBS PMS protocol to optimize sample input for five species of coral: P. lutea, M. capitata, P. acuta, P. meandrina, P. asterodies.
Both trials done in 2 days - day 1 ends with the bisulfite conversion step, left in the 4C fridge for less than 20 hours and then started again the next day. This could probably be done in 1 day in the future now that I have done this protocol multiple times.
Trial 1: 20201015 and 20201026
DNA input: 1 ng. Dilutions done by taking 2 uL of DNA + (1-Qubit value)x2 10 uM Tris HCl.
BR DNA Standard 1: 200.79
BR DNA Standard 2: 22,255.60
| Coral_ID | Species | Qubit (ng/uL) | Gel Pass? | DNA for dilution (ul) | 10 mM Tris for dilution (ul) | Qubit (ng/uL) post PMS | TapeStation pass? | Notes |
|---|---|---|---|---|---|---|---|---|
| P7 | Pocillopora meandrina | 75.6 | Not good | 2 | 149.2 | Too low | Peaks visible | Hawaii - Eva extracted this with HMW kit and sent it in the recent dry shipper |
| P3 | Porites lutea | 136.5 | Not good | 2 | 271 | Too low | Peaks visible | Hawaii - Eva extracted this with HMW kit and sent it in the recent dry shipper |
| P1 | Porites astreoides | 8.27 | Great | 2 | 14.54 | 2.08 | Peaks visible | genome coral - Maggie extracted this with HMW kit |
| Date | Sample | i5 index # | i7 index # |
|---|---|---|---|
| 20201025 | P1 - Past | 1 | 1 |
| 20201025 | P3 - P lutea | 2 | 2 |
| 20201025 | P7 - Poc mea | 3 | 3 |
Calculations for mixes:
Amplification with Prep-Amp Primers
3 samples + 0.15 (for 5% error) = 3.15
Priming Master Mix (PMM):
6.3 uL of 5X PrepAmp Buffer (2 uL * 3.15 samples = 6.3)
3.15 uL 40 uM PrepAmp Primer (1 uL * 3.15 samples = 3.15)
PrepAmp Master Mix (PAMM):
3.15 uL of 5X PrepAmp Buffer (1 uL * 3.15 samples = 3.15)
11.81 uL PrepAmp Pre Mix (3.75 uL * 3.15 samples = 11.81)
0.95 uL PrepAmp Polymerase (0.3 uL * 3.15 = 0.95)
“Diluted” PrepAmp Polymerase mix
0.9 uL PrepAmp Polymerase (0.3 uL * 3 samples = 0.9)
6.3 uL DNA Elution Buffer (0.2 uL * 3.15 = 6.3)
*The above calculation should be 0.63 uL and 0.99 uL, unclear if that was a written mistake or a mistake made during the protocol. This mix could have been far too diluted for the protocol, leading to less enzymatic activity and smaller output.**
1st Amplification Master Mix (AMM)
39.38 uL Library Amp Mix (2X) (12.5 uL * 3.15 samples = 39.375)
3.15 uL Library Amp Primers (10 uM) (1 uL * 3.15 samples = 3.15)
20201026 Trial 1 TapeStation Report
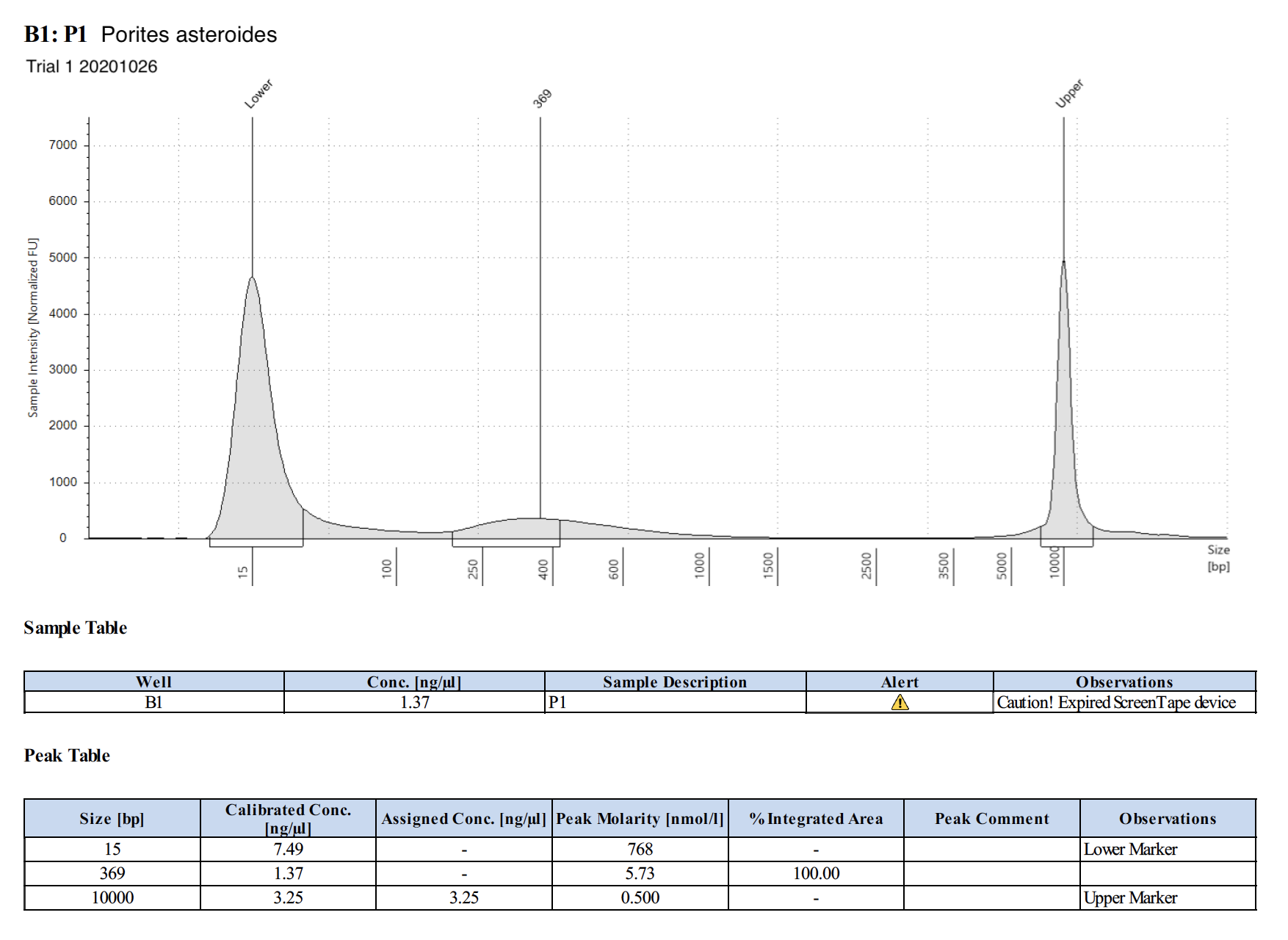
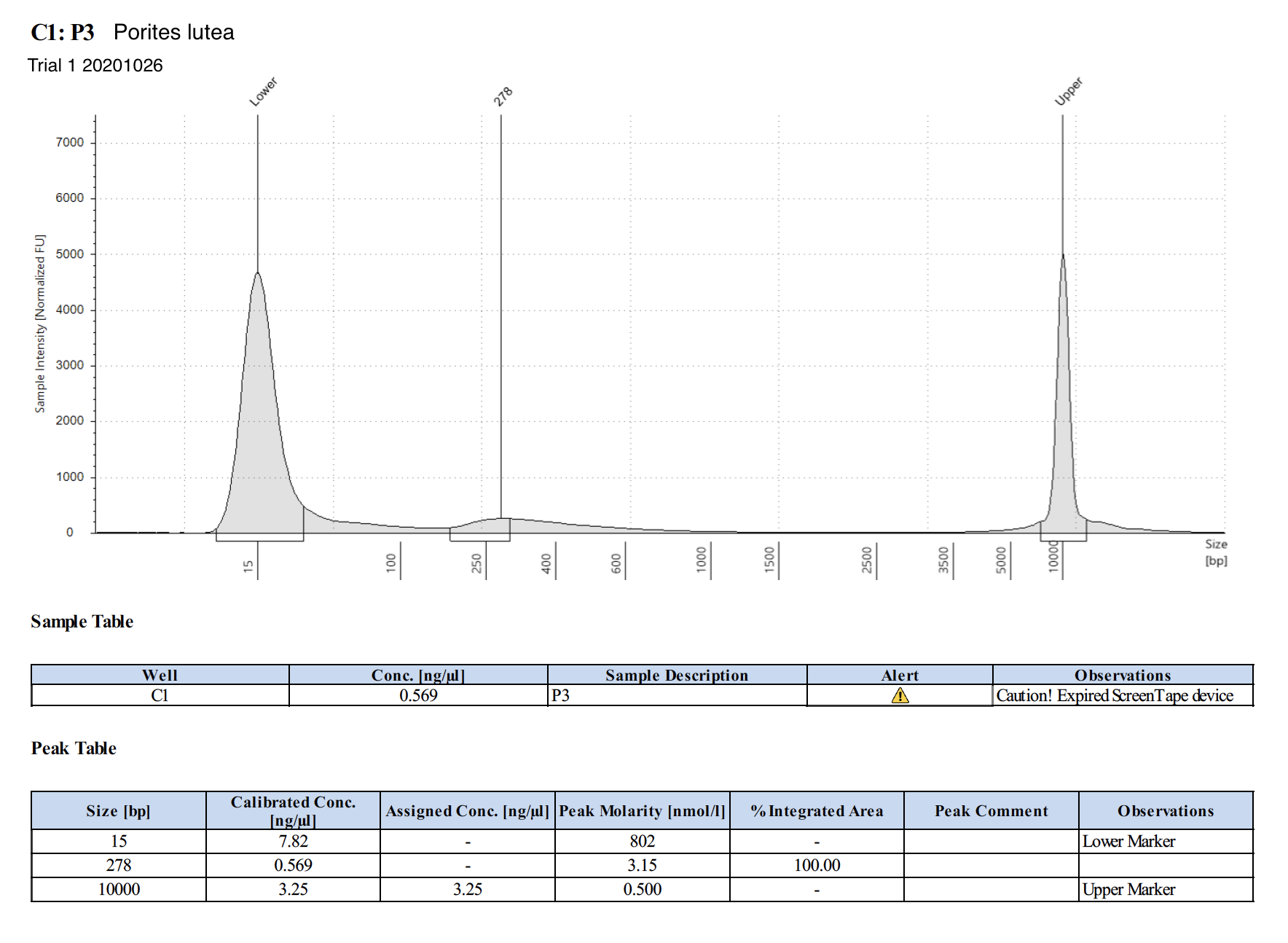
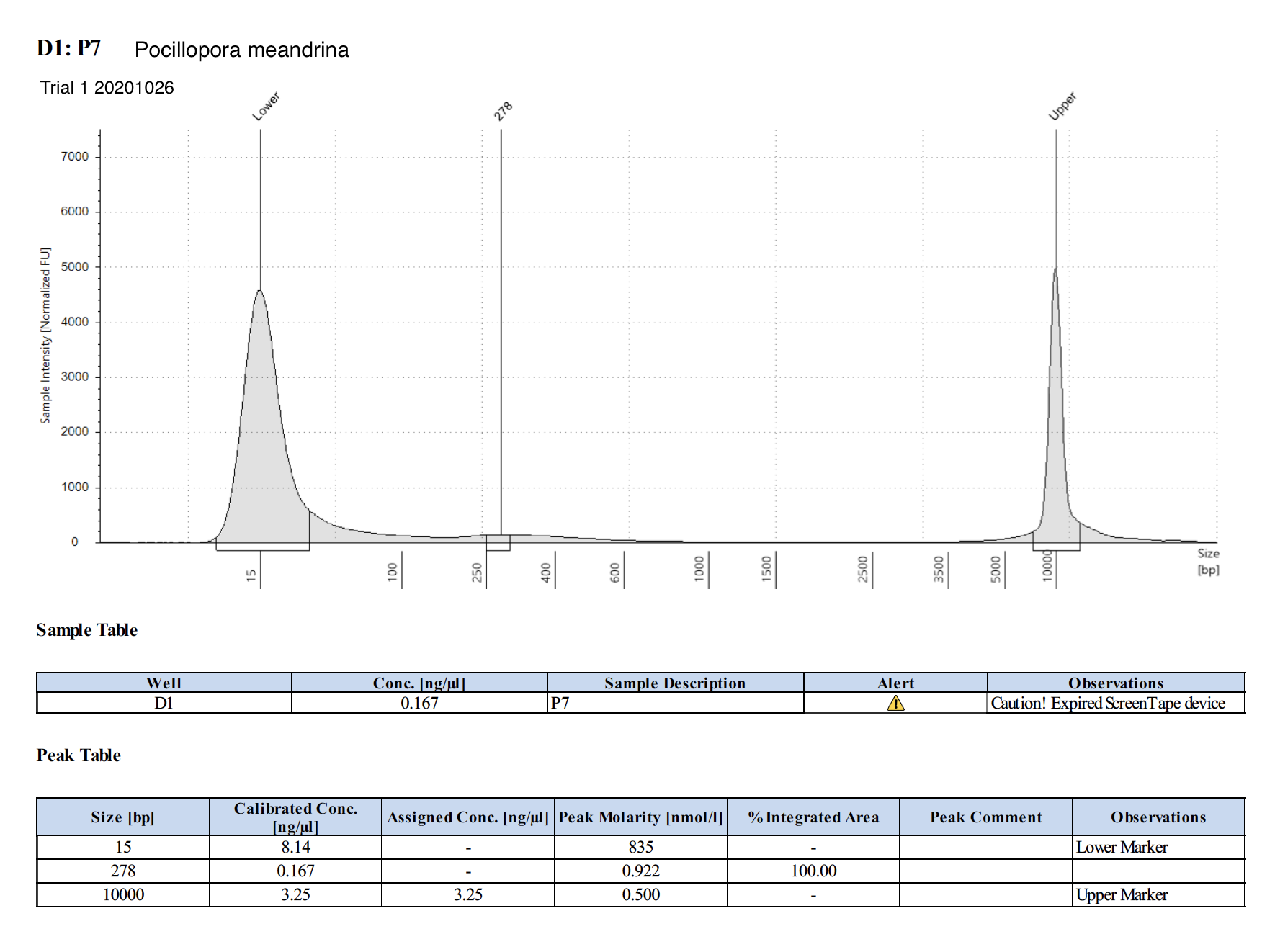
Trial 2: 20201110 and 20201111
DNA input: 10 ng. 10 ng/uL calculations done by 100/Qubit value uL DNA for dilution and 10-DNA value for 10 uM Tris HCl volume.
BR DNA Standard 1: 180.28
BR DNA Standard 2: 19,255.88
| Coral_ID | Species | Qubit (ng/uL) | Gel Pass? | DNA needed (uL) | 10 mM Tris HCl (uL) | Qubit (ng/uL) post PMS | TapeStation pass? | Notes |
|---|---|---|---|---|---|---|---|---|
| P3 | Pocillopora meandrina | 54.75 | Good | 1.83 | 8.17 | 3.81 | Peaks visible | Maggie extracted HMW from 20201109 |
| P1 | Porites astreoides | 8.27 | Good | Too low to dilute; just take 1 uL of original tube | Too low | Peaks visible | genome coral - Maggie extracted this with HMW kit | |
| 2860 | Montipora capitata | 43.5 | Good | 2.30 | 7.70 | 2.58 | Peaks visible | Emma extracted - Holo Int |
| 2878 | Pocillopora acuta | 48.1 | Fair | 2.08 | 7.92 | 3.81 | Peaks visible | Emma extracted - Holo Int |
Index Key
| Sample Name/Pool Name* | Library Name* | i7 Index Name* | i7 Index Sequence* | i5 Index Name* | i5 Index Sequence* |
|---|---|---|---|---|---|
| Mcap_2860 | Mcap_2860 | i7_UDI0001 | AACCGCGG | i5_UDI0001 | AGCGCTAG |
| Pact_2878 | Pact_2878 | i7_UDI0002 | GGTTATAA | i5_UDI0002 | GATATCGA |
| Past_P1 | Past_P1 | i7_UDI0003 | CCAAGTCC | i5_UDI0003 | CGCAGACG |
| Pmean_P3 | Pmean_P3 | i7_UDI0004 | TTGGACTT | i5_UDI0004 | TATGAGTA |
I started my samples at 1 and 2 in case this trial worked and I could start the rest of my samples at 3.
Mix Calculations
Amplification with Prep-Amp Primers
4 samples + 0.2 (for 5% error) = 4.2
Priming Master Mix (PMM):
8.4 uL of 5X PrepAmp Buffer (2 uL * 4.2 samples = 8.4)
4.2 uL 40 uM PrepAmp Primer (1 uL * 4.2 samples = 4.2)
PrepAmp Master Mix (PAMM):
4.2 uL of 5X PrepAmp Buffer (1 uL * 4.2 samples = 4.2)
15.75 uL PrepAmp Pre Mix (3.75 uL * 4.2 samples = 15.75)
1.26 uL PrepAmp Polymerase (0.3 uL * 4.2 = 1.26)
“Diluted” PrepAmp Polymerase mix
1.26 uL PrepAmp Polymerase (0.3 uL * 4.2 samples = 1.26)
0.84 uL DNA Elution Buffer (0.2 uL * 4.2 = 0.84)
1st Amplification Master Mix (AMM)
52.5 uL Library Amp Mix (2X) (12.5 uL * 4.2 samples = 52.5)
4.2 uL Library Amp Primers (10 uM) (1 uL * 4.2 samples = 4.2)
20201111 Trial 2 TapeStation Report
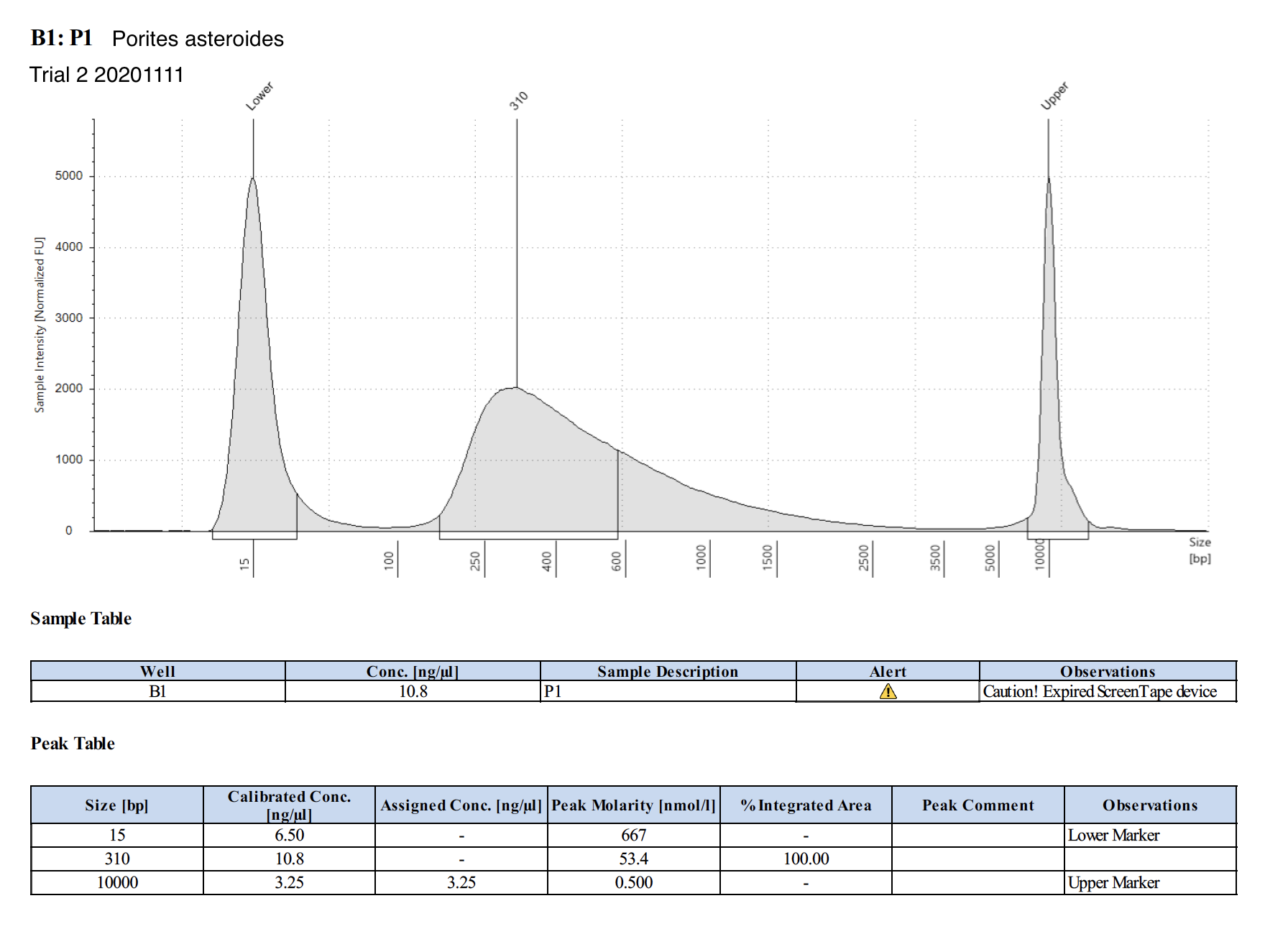
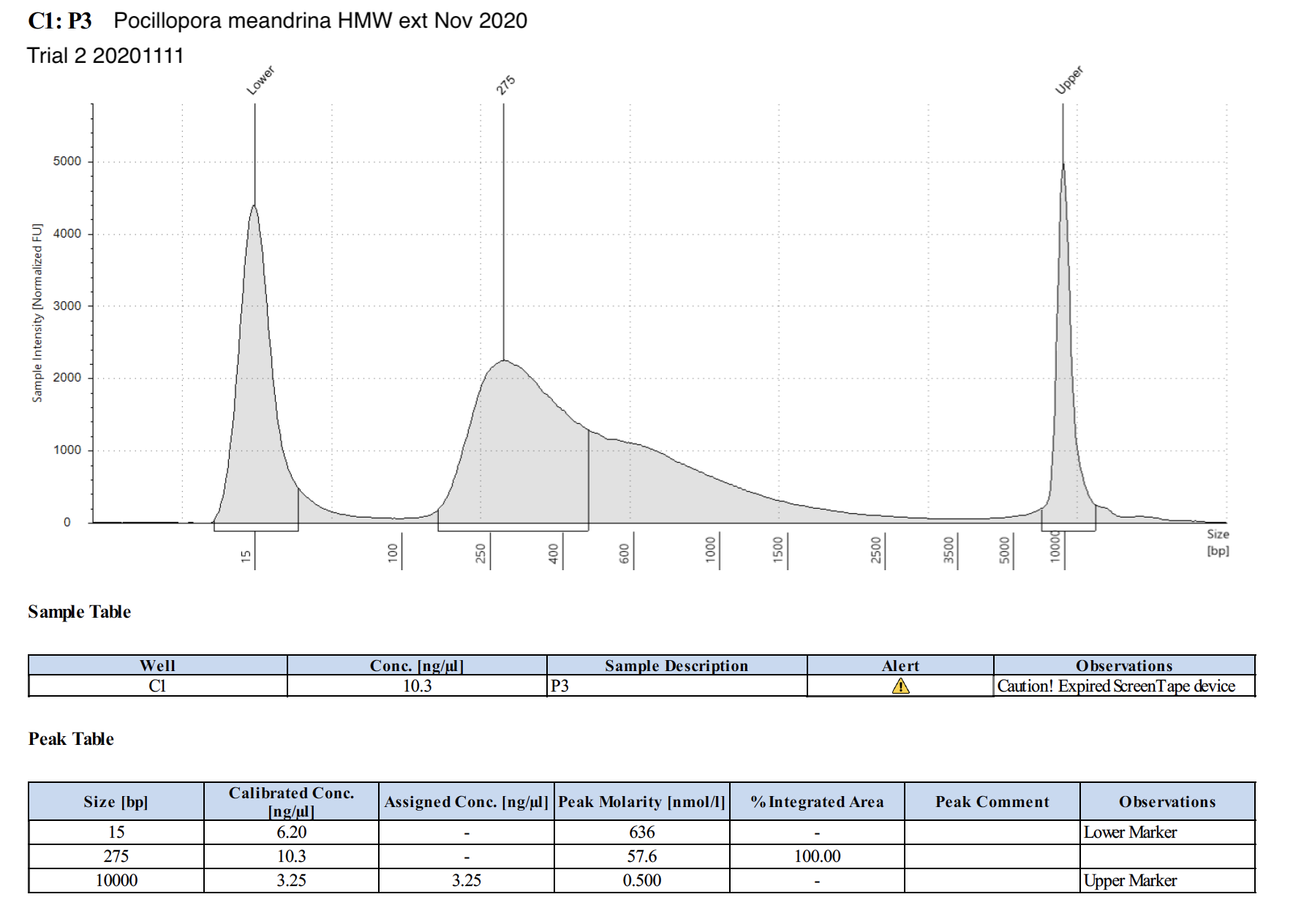
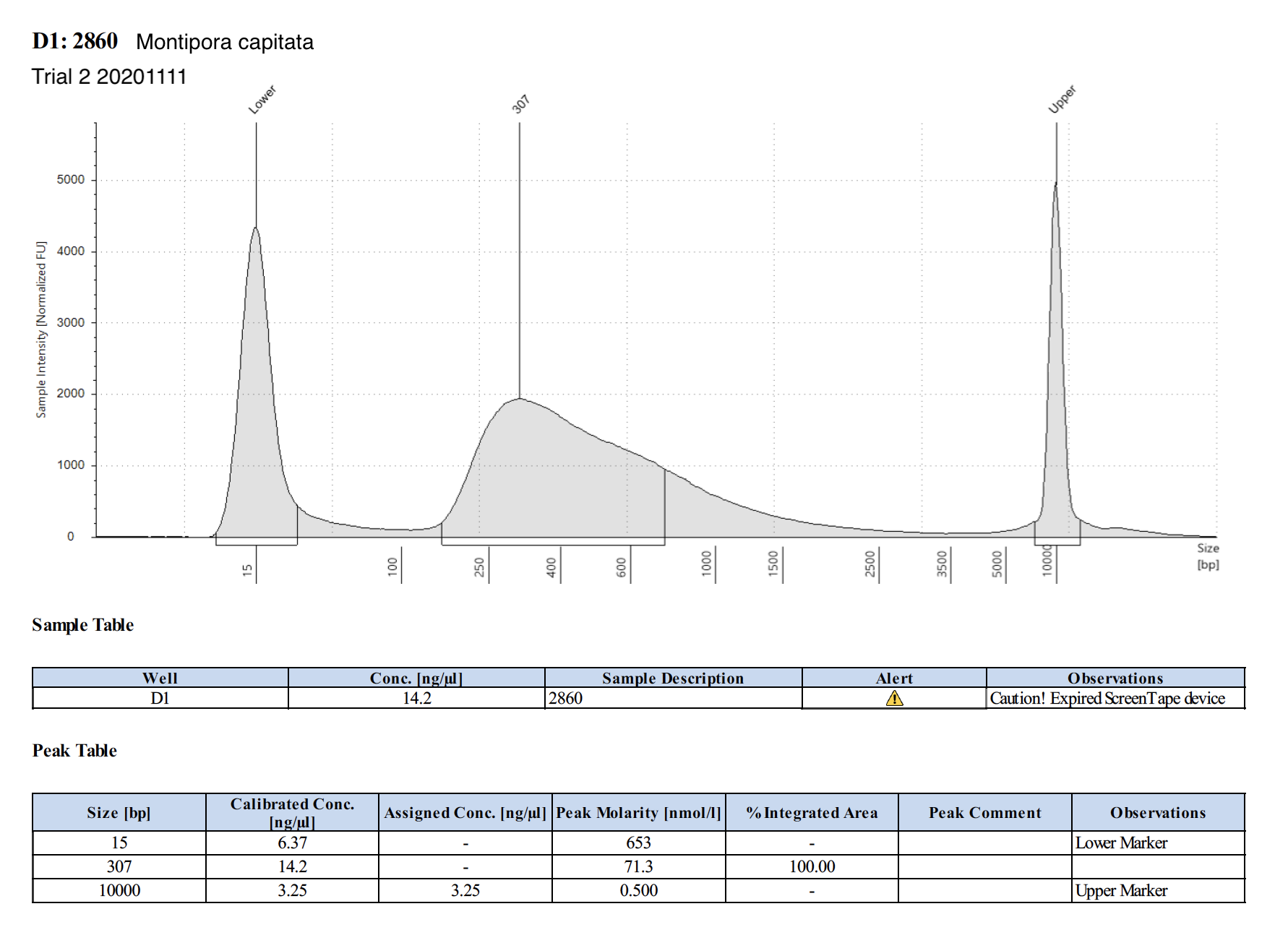
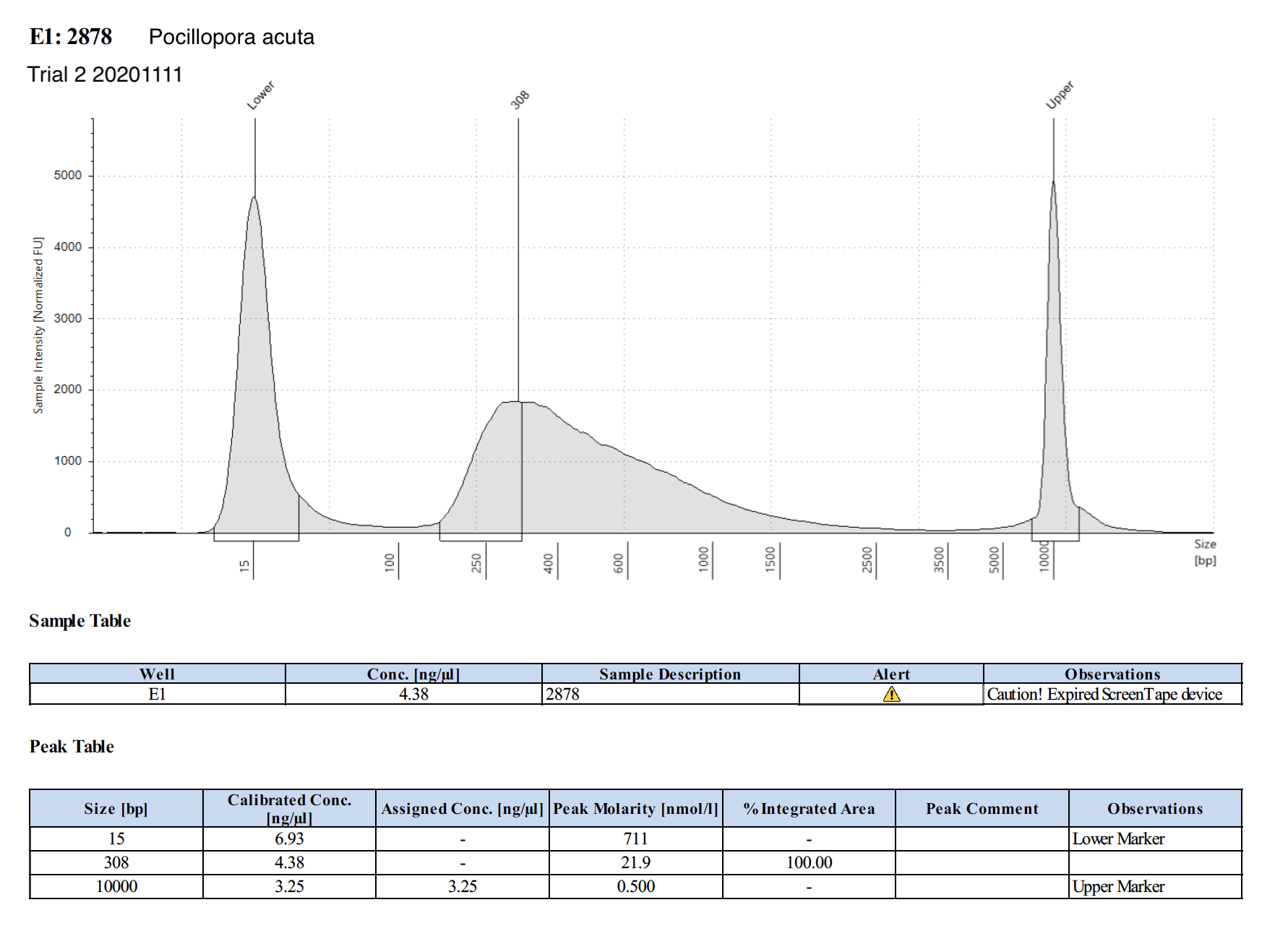
The concentrations are much higher on the TapeStation than Qubit values. Which one do we trust more? The output of WGBS PMS is only 15 uL max, and I used 3 uL for tapestation and qubit (prepped qubit twice). I’m hesitant to use more to re-qubit unless we have to.
Submission details (20201117_30-445814566_Strand_WGBS):
| Sample # | General Information | Amount of Material | Miscellaneous | Special Comments | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SeqFile-Name | Sample/Pool Name* | Number of Libraries in Pool* | Starting Material Type* | Species/Strain* | Sample Buffer* | Total Amount* | Volume (µL)* | Conc. (nM or ng/uL)* | Purification Method | Library Prep Kit Used* | ||
| 1 | ES1 | Mcap_2860 | 1 | DNA | Porites astreoides | DNA elution buffer | 25.8 | 10 | 2.58 | KAPA Pure Beads | Zymo Pico Methyl-Seq Library Prep Kit | |
| 2 | ES2 | Pact_2878 | 1 | DNA | Pocillopora meandrina | DNA elution buffer | 38.1 | 10 | 3.81 | KAPA Pure Beads | Zymo Pico Methyl-Seq Library Prep Kit | |
| 3 | ES3 | Past_P1 | 1 | DNA | Montipora capitata | DNA elution buffer | 10 | 10 | 1 | KAPA Pure Beads | Zymo Pico Methyl-Seq Library Prep Kit | |
| 4 | ES4 | Pmean_P3 | 1 | DNA | Pocillopora acuta | DNA elution buffer | 38.1 | 10 | 3.81 | KAPA Pure Beads | Zymo Pico Methyl-Seq Library Prep Kit |
