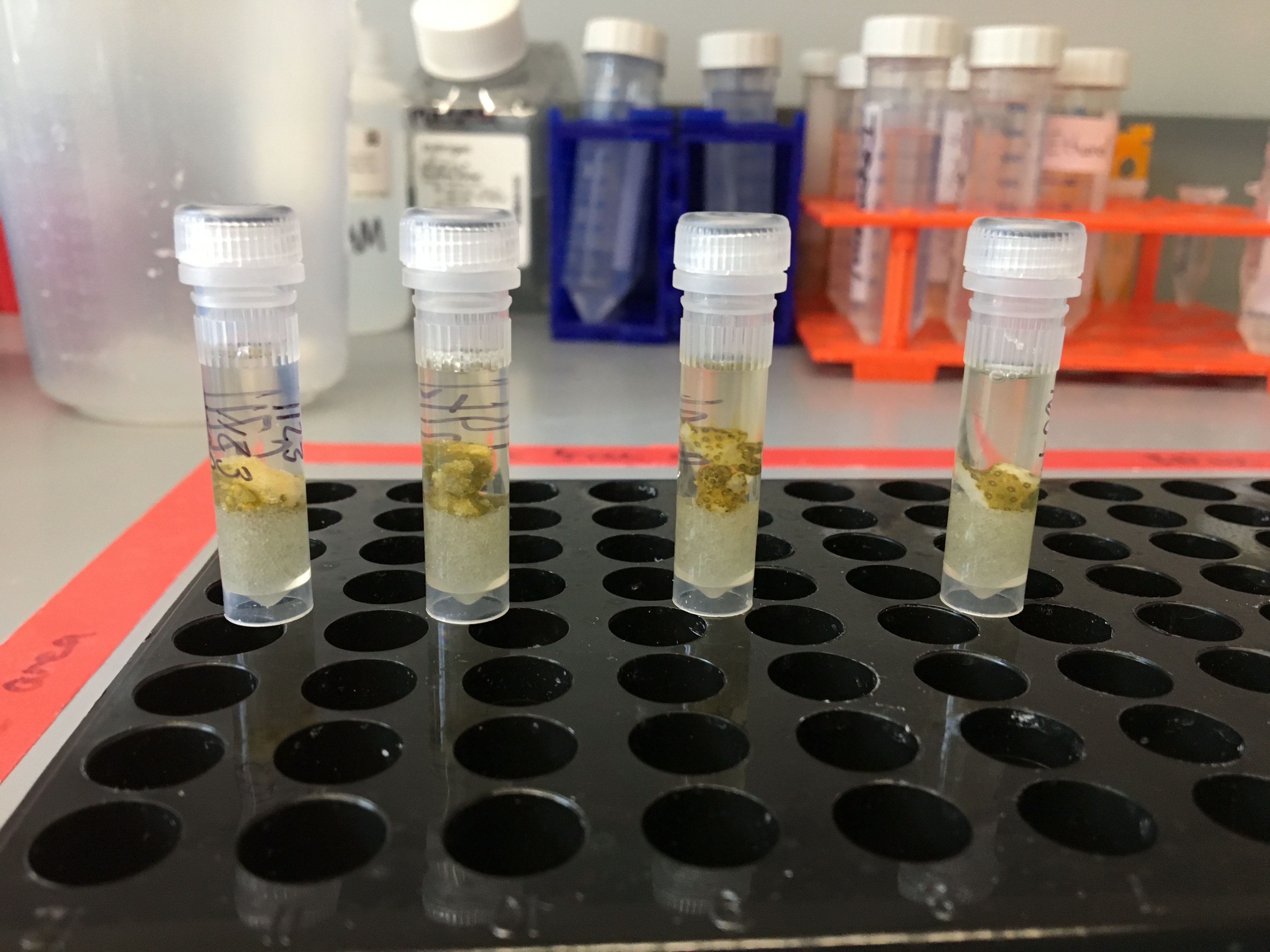
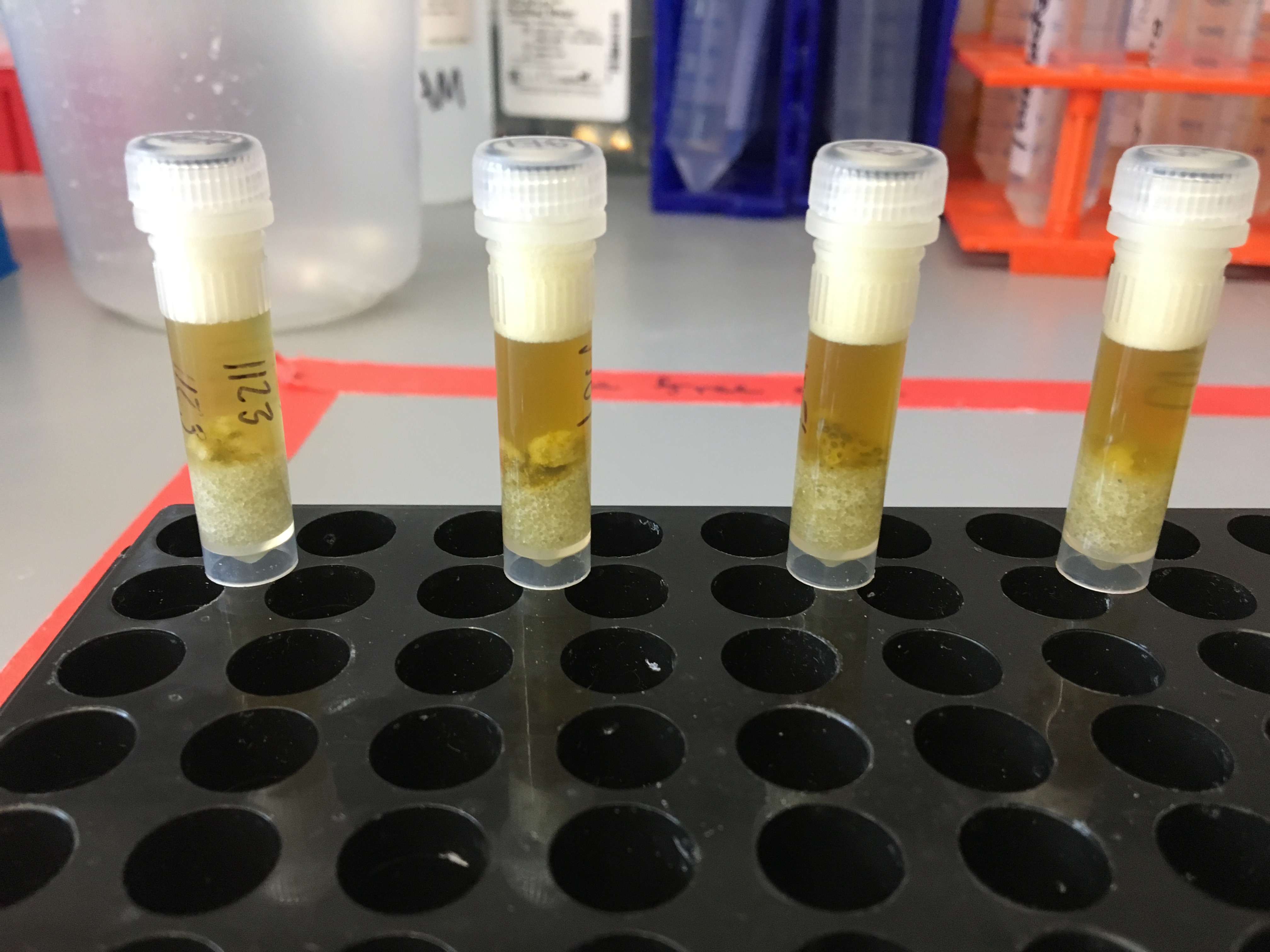
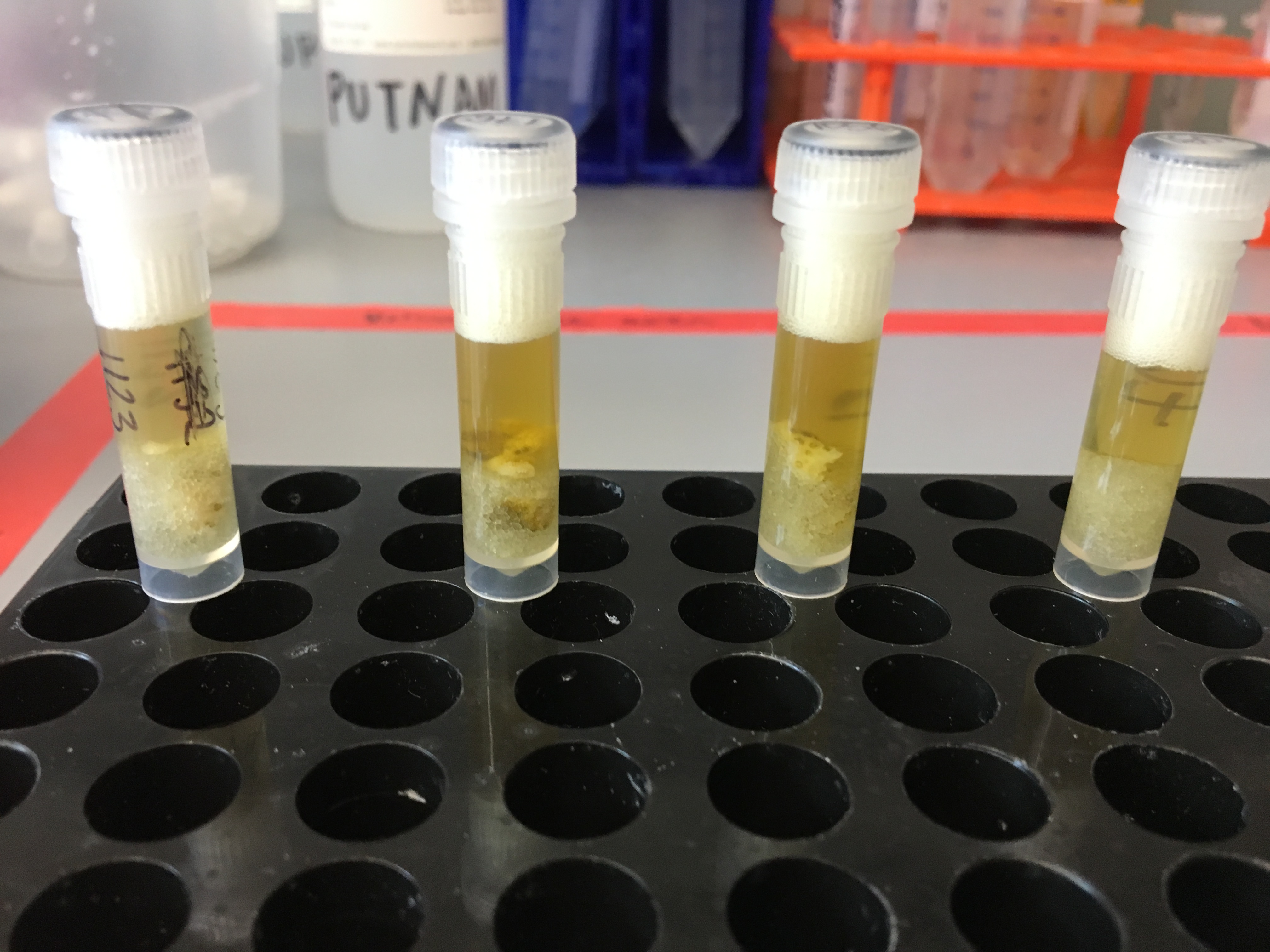
Testing Soft and Hard Homogenization Round 4
20190716 E. Strand
Testing the soft and hard homogenization protocol on extra molecular samples from the December recovery time period of the Holobiont Integration project. 2 M. capitata and 2 P. acuta fragments were randomly chosen from ATAC treatment 20181215. These four fragments were used on 20190715 and 20190716.
| Extraction # | Coral ID | Species | Homogenization |
|---|---|---|---|
| 1 | 1123 | Montipora | Soft |
| 2 | 1123 | Montipora | Hard |
| 3 | 1769 | Montipora | Soft |
| 4 | 1769 | Montipora | Hard |
| 5 | 1056 | Pocillopora | Soft |
| 6 | 1056 | Pocillopora | Hard |
| 7 | 1607 | Pocillopora | Soft |
| 8 | 1607 | Pocillopora | Hard |
Soft and Hard homogenization, and DNA/RNA Extractions followed this protocol. General Zymo Duet RNA DNA Extraction protocol found here.
With the following changes:
- No heat to lyse step to test if the heating is contributing to the RNA degradation.
- Just 1 fragment clipping instead of 2-3 clippings to test if the RNA/DNA shield is overloaded with more than one clipping.
- Diluted soft homogenization supernatant with 500 µl of RNA/DNA shield accidentally. Results suggest that this didn’t impact the quality, but may have diluted quantity.
Before homogenization steps:
After soft homogenization:
After hard homogenization:
Extraction steps: start 10:00 end 12:06. 8 samples takes about two hours.
1056 soft DNA final tube contains ~70-80 µl instead of 90 µl.
DNAse I Master Mix Calculations:
- 75 µl x 8 samples = 600 µl buffer
- 5 µl DNase I x 8 samples = 40 µl DNase I enzyme
Qubit Results:
Master Mix Calculations:
- 8 samples + 2 standards + 0.2% error = 10.2 µl Quant-IT Reagent
- 199 * 10.2 = 2,029.8 µl Qubit buffer
| DNA (ng/μl) | |||
|---|---|---|---|
| Standard 1 | 176.27 | ||
| Standard 2 | 20135.99 | ||
| 1 | 1132 Mcap soft | 20.8 | 20.8 |
| 2 | 1132 Mcap hard | 20.8 | 20.6 |
| 3 | 1769 Mcap soft | 27 | 26.8 |
| 4 | 1769 Mcap hard | 28 | 28 |
| 5 | 1056 Pacuta soft | 39.4 | 39.4 |
| 6 | 1056 Pacuta hard | 54 | 53.8 |
| 7 | 1607 Pacuta soft | 18.6 | 18.5 |
| 8 | 1607 Pacuta hard | 17.4 | 17.3 |
| RNA (ng/μl) | |||
|---|---|---|---|
| Standard 1 | 393.16 | ||
| Standard 2 | 10592.52 | ||
| 1 | 1132 Mcap soft | 11.8 | 11.8 |
| 2 | 1132 Mcap hard | 10.6 | 10.6 |
| 3 | 1769 Mcap soft | 16.6 | 16.6 |
| 4 | 1769 Mcap hard | 20.4 | 20.4 |
| 5 | 1056 Pacuta soft | 34.2 | 34.2 |
| 6 | 1056 Pacuta hard | 47.2 | 47.2 |
| 7 | 1607 Pacuta soft | 27.8 | 27.8 |
| 8 | 1607 Pacuta hard | 22.8 | 22.8 |
If the quantity is below 10 ng/μl in the soft homogenization, then re-extract DNA and RNA to allow for enough DNA/RNA for methylation and transcriptomic protocols/sequencing.
- 100V for 45 minutes, start 12:47 end 13:32
- 75 μl TAE buffer + 0.75 g agarose + 5 μl of Biotium gel RED gel stain to make a 1.5% gel.
- 5 μl samples + 1 μl 6x purple loading dye
- 4 μl GeneRuler 1 kb Plus DNA ladder + 1 μl 6x purple loading dye
Gel order: Ladder, 1-8
Ordered Biotium gel GREEN gel stain to use in the future instead of Biotium gel RED gel stain. Gel GREEN stain has been brighter in gels before for labmates.
TapeStation Results:
- Thermocycler (rna denature program): 3 minutes at 72 °C, 2 minutes at 4 °C, hold at 4 °C.
- TapeStation start 14:10 end 14:20
- Ladder: Agilent RNA screentape ladder
| Extraction ID | Coral ID | RIN^e |
|---|---|---|
| 1 | 1132 Mcap soft | 6.1 |
| 2 | 1132 Mcap hard | ** |
| 3 | 1769 Mcap soft | 7.7 |
| 4 | 1769 Mcap hard | 8.2 |
| 5 | 1056 Pacuta soft | 6.9 |
| 6 | 1056 Pacuta hard | 7.1 |
| 7 | 1607 Pacuta soft | 6.3 |
| 8 | 1607 Pacuta hard | 6.0 |
** Concentration too low for TapeStation to detect. But two small peaks are still visible in both reports. TapeStation done twice because samples were out of range. Not necessary in the future since a Qubit result of equal to or less than 10 ng/µl will likely read as too low of a concentration.
Link to the first 20190716 report
Link to the second 20190716 report
Link to Agilent 4200 TapeStation System
Future changes for next “round” of testing this protocol:
- Change hard homogenization in Tissue Lyser step to 1 minute instead of 30 seconds.