Testing Soft and Hard Homogenization Round 3
Soft and Hard Homogenization Protocol Testing Round 3
20190715 E. Strand
Testing the soft and hard homogenization protocol on extra molecular samples from the December recovery time period of the Holobiont Integration project. 2 M. capitata and 2 P. acuta fragments were randomly chosen from ATAC treatment 20181215.
| Extraction # | Coral ID | Species | Homogenization |
|---|---|---|---|
| 1 | 1123 | Montipora | Soft |
| 2 | 1123 | Montipora | Hard |
| 3 | 1769 | Montipora | Soft |
| 4 | 1769 | Montipora | Hard |
| 5 | 1056 | Pocillopora | Soft |
| 6 | 1056 | Pocillopora | Hard |
| 7 | 1607 | Pocillopora | Soft |
| 8 | 1607 | Pocillopora | Hard |
Fragment #1607 had algae covering the top of the fragment. Pieces were broken off at the bottom and used for the following extraction protocol.
Soft and Hard homogenization, and DNA/RNA Extractions followed this protocol. General Zymo Duet RNA DNA Extraction protocol found here.
2-3 fragment pieces were clipped per coral. The following photo depicts the fragment pieces in RNA DNA shield before soft and hard homogenization steps.
Pocillopora were “soft-homogenized” for 1 minute and Montipora for 2 minutes in a vortex. Both species were “hard-homogenized” for 30 seconds at 20 Hz in the Qiagen Tissue Lyser: Handbook.
Extraction notes:
- New Proteinase K made halfway through steps, new Proteinase K used for samples #6-8.
- Heat to lyse step start 10:30 end 12:00
- Extraction steps start 12:02 end ~14:00
Fragment pieces after the soft homogenization:
Fragment pieces after the hard homogenization:
DNA Master Mix Calculations:
- 75 μl DNA Digestion Buffer x 8 samples = 600 μl buffer
- 5 μl DNase I x 8 samples = 40 μl DNase I enzyme
Qubit Master Mix Calculations:
- 8 samples + 2 standards + 0.2% error = 10.2 μl Quant-IT reagent
- 199 * 10.2 = 2,029.8 μl buffer
Qubit (ng/μl) Results:
| DNA | |||
|---|---|---|---|
| Standard 1 | 239.46 | ||
| Standard 2 | 23085.86 | ||
| 1 | 1132 Mcap soft | 12.5 | 12.4 |
| 2 | 1132 Mcap hard | too low | too low |
| 3 | 1769 Mcap soft | 5.32 | 5.24 |
| 4 | 1769 Mcap hard | 4 | 3.9 |
| 5 | 1056 Pacuta soft | 9.74 | 9.5 |
| 6 | 1056 Pacuta hard | too low | too low |
| 7 | 1607 Pacuta soft | 4.18 | 4.12 |
| 8 | 1607 Pacuta hard | too low | too low |
| RNA | |||
|---|---|---|---|
| Standard 1 | 390.27 | ||
| Standard 2 | 10287.94 | ||
| 1 | 1132 Mcap soft | 38.4 | 38.2 |
| 2 | 1132 Mcap hard | 18.2 | 18.2 |
| 3 | 1769 Mcap soft | 44.6 | 44.4 |
| 4 | 1769 Mcap hard | 26 | 26.2 |
| 5 | 1056 Pacuta soft | 73.8 | 73.6 |
| 6 | 1056 Pacuta hard | 44.2 | 44 |
| 7 | 1607 Pacuta soft | 79 | 79 |
| 8 | 1607 Pacuta hard | 45.4 | 45.4 |
The value for DNA Standard 2 looks too high. Standards and Qubit re-done.
Qubit (ng/μl) round 2 results:
| DNA | |||
|---|---|---|---|
| Standard 1 | 152.27 | ||
| Standard 2 | 10492.87 | ||
| 1 | 1132 Mcap soft | 19 | 18.7 |
| 2 | 1132 Mcap hard | 6.8 | 6.74 |
| 3 | 1769 Mcap soft | 11 | 10.9 |
| 4 | 1769 Mcap hard | 9.24 | 9.12 |
| 5 | 1056 Pacuta soft | 17.9 | 17.5 |
| 6 | 1056 Pacuta hard | 2.96 | 2.72 |
| 7 | 1607 Pacuta soft | 8.62 | 8.42 |
| 8 | 1607 Pacuta hard | 3.18 | 3.22 |
| RNA | |||
|---|---|---|---|
| Standard 1 | 393.13 | ||
| Standard 2 | 10255.01 | ||
| 1 | 1132 Mcap soft | 41.2 | 41.2 |
| 2 | 1132 Mcap hard | 26.4 | 26.2 |
| 3 | 1769 Mcap soft | 48.2 | 48.2 |
| 4 | 1769 Mcap hard | 26 | 25.8 |
| 5 | 1056 Pacuta soft | 77 | 77 |
| 6 | 1056 Pacuta hard | 51 | 51 |
| 7 | 1607 Pacuta soft | 83.4 | 83.2 |
| 8 | 1607 Pacuta hard | 48 | 48 |
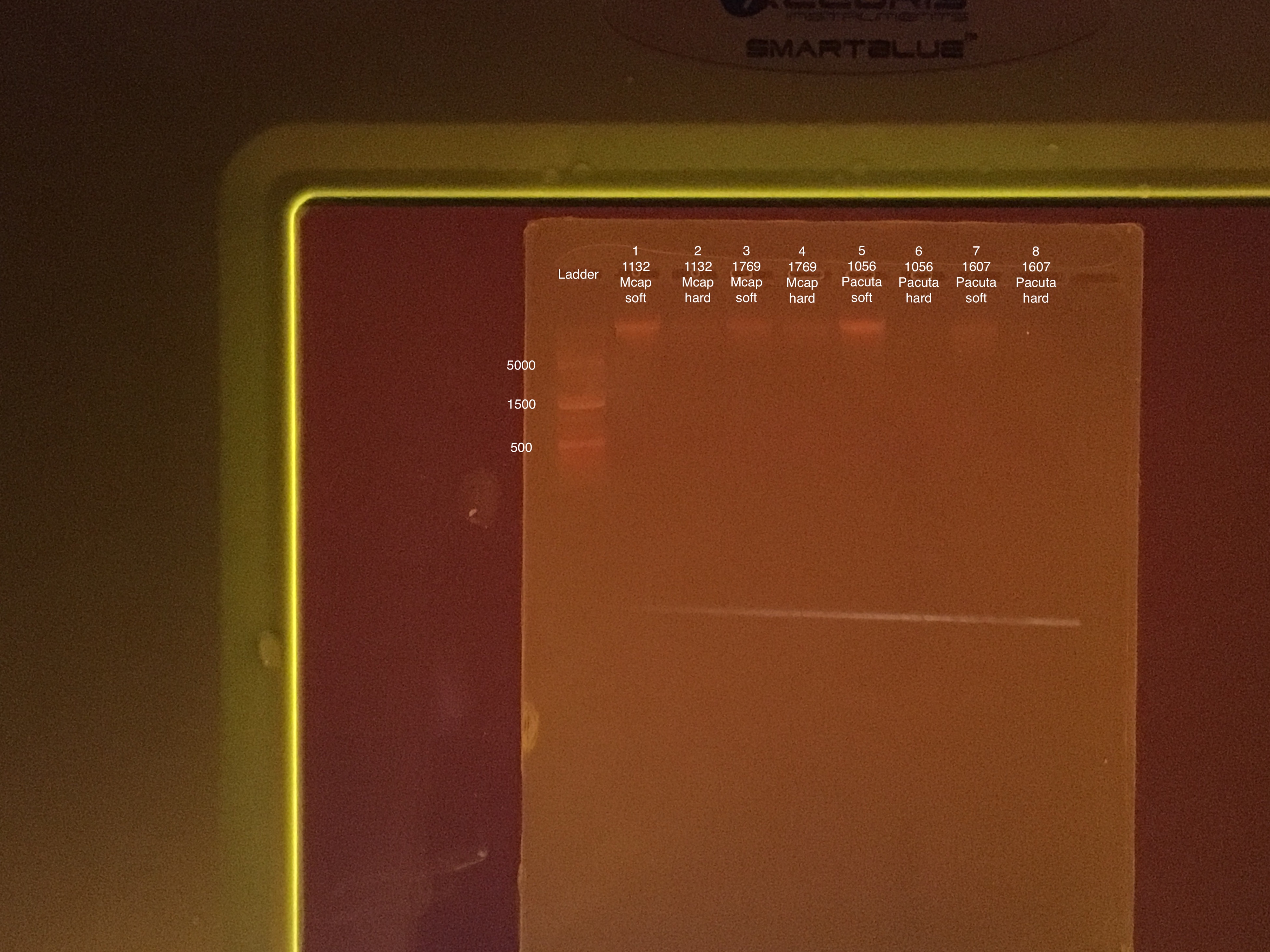
- 100V for 45 minutes, start 15:52 end 16:27
- 75 μl TAE buffer + 0.75 g agarose + 5 μl of Biotium gel RED gel stain to make a 1.5% gel
- 5 μl samples + 1 μl 6x purple loading dye
- 4 μl GeneRuler 1 kb Plus DNA ladder + 1 μl 6x purple loading dye
Sample order: Ladder, #1-8
TapeStation Results:
| Extraction ID | Coral ID | RIN^e |
|---|---|---|
| 1 | 1132 Mcap soft | 5.8 |
| 2 | 1132 Mcap hard | 4.8 |
| 3 | 1769 Mcap soft | 6.8 |
| 4 | 1769 Mcap hard | 5.3 |
| 5 | 1056 Pacuta soft | 6.1 |
| 6 | 1056 Pacuta hard | 5 |
| 7 | 1607 Pacuta soft | 5 |
| 8 | 1607 Pacuta hard | 4.3 |
Link to the full 20190715 report Link to Agilent 4200 TapeStation System
- Thermocycler (rna denature program): 3 minutes at 72 °C, 2 minutes at 4 °C, hold at 4 °C.
- TapeStation start 16:17 end 16:27
- Ladder: Agilent RNA screentape ladder