Testing Soft and Hard Homogenization Round 2
Soft and Hard Homogenization Protocol Testing Round 2
20190610 E.S.
We chose one M. capitata and one P. acuta of the four from Testing Round 1 to test the vortex timing in the soft homogenization step. During the first round of protocol testing, the tissue on the P. acuta fragments came off much quicker than the M. capitata and therefore might need a shorter vortex time during the soft homogenization.
Soft and Hard homogenization, and DNA/RNA Extractions followed this protocol. General Zymo Duet RNA DNA Extraction protocol found here.
Modified from original Zymo Duet RNA/DNA Extraction protocol
Samples used:
| ORIGIN | Site.Name | POINT | SITE.ID | Species | COLLECT.DATE | PLUG.ID | TANK# | TREATMENT | ANALYSIS | TIME | TIME POINT | SAMPLING.DATE | Dead.Alive | NOTES | Sample Location | Shipment Date | Lab Work | Lab Work by | Lab Work Date |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SITE 4 | Reef.11.13 | 6 | P60 | Pacuta | 20180907 | 1431 | 6 | ATAC | Molecular | 11:24 | 11 | 20181215 | Alive | In Tank 6 20181118; off plug | In Transit to URI | 20190206 | Soft and hard homogenization | ES | 20190606; 20190610 |
| SITE 1 | Lilipuna.Fringe | 6 | M54 | Mcapitata | 20180908 | 1591 | 8 | ATAC | Molecular | 10:12 | 11 | 20181215 | Alive | In Tank 8 20181118 | In Transit to URI | 20190206 | Soft and hard homogenization | ES | 20190606; 20190610 |
| Extraction # | Coral ID | Species | Soft homogenization time |
|---|---|---|---|
| 1 | 1431 | P. acuta | 30 seconds |
| 2 | 1431 | P. acuta | 1 minute |
| 3 | 1431 | P. acuta | 1.5 minutes |
| 4 | 1431 | P. acuta | 2 minutes |
| 5 | 1591 | M. capitata | 30 seconds |
| 6 | 1591 | M. capitata | 1 minute |
| 7 | 1591 | M. capitata | 1.5 minutes |
| 8 | 1591 | M. capitata | 2 minutes |
General extraction notes:
- Clipped coral fragments into 4-5 pieces small enough to fit in a 1.5 microcentrifuge tube.
- Tubes #2 and #4 look a lot paler than #1 and #3 (all 1431 P. acuta). This could be because of the size of cut pieces and position tube. P. acuta seems to be more sensitive than the M. capitata pieces.
- Tubes #5-8 coloring looks similar across all four tubes. Tube #5 looks like it still has the most tissue on the skeleton after vortexing. Which makes sense it was vortexed for a shorter amount of time.
- Hard homogenization was done with the vortex for 30 seconds at a time until most of the tissue was off the skeleton. We were out of bead bug tubes and beads, and the Tissue Lyster is still broken. Hard homogenization went for 1.5 minutes. All tubes had the same amount of time for hard homogenization.
- Tubes #5 and #6 were very mucus-y and harder to pipette. The tissue doesn’t seem to be broken down enough.
- After hard and soft homogenizations, I would predict that M. capitata needs at least 1.5 minutes - 2 minutes and P. acuta needs around 1 minute. But the size and position of P. acuta fragment pieces in the tube seem to affect the time needed in soft homogenization more than the M. capitata fragment pieces.
- 300 uL of sample was taken for extraction. 30 uL Proteinase K Buffer and 15 uL Proteinase K were added to the sample before heating step. See linked Zymo protocol above for details on ratios.
- Incubation started at 10:51, end at 12:20
- Extractions ended at 3:45, qubit at 4:00
Calculations:
DNAse I master mix:
- 75 uL x 16 samples (8 original samples x 2 for soft and hard) = 1,200 buffer
- 5 uL x 16 samples (see above) = 80 uL DNAse I
Qubit master mix:
Once for DNA and once for RNA:
- 16 samples (see above) + 2 standards + 0.2% error = 18.2 uL of Quant-IT reagent
- 199 x 18.2 = 3,621.8 uL of buffer
Qubit Results (Protocol)
| DNA | |||||
|---|---|---|---|---|---|
| Coral ID | Extraction ID | Extraction type | Trial 1 | Trial 2 | Avg (ng/uL) |
| 1431 | 1 | soft | 145 | 145 | 145 |
| 1431 | 1 | hard | 73.8 | 73.4 | 73.6 |
| 1431 | 2 | soft | 136 | 136 | 136 |
| 1431 | 2 | hard | 40.6 | 40.4 | 40.5 |
| 1431 | 3 | soft | 60.4 | 60.2 | 60.3 |
| 1431 | 3 | hard | 80 | 79.6 | 79.8 |
| 1431 | 4 | soft | 154 | 153 | 153.5 |
| 1431 | 4 | hard | 58 | 57.8 | 57.9 |
| 1591 | 5 | soft | 50.8 | 50.4 | 50.6 |
| 1591 | 5 | hard | 32.8 | 32.6 | 32.7 |
| 1591 | 6 | soft | 34.6 | 34.4 | 34.5 |
| 1591 | 6 | hard | 57.6 | 57.4 | 57.5 |
| 1591 | 7 | soft | 66.4 | 66.2 | 66.3 |
| 1591 | 7 | hard | 66 | 65.6 | 65.8 |
| 1591 | 8 | soft | 130 | 129 | 129.5 |
| 1591 | 8 | hard | 55.8 | 55.6 | 55.7 |
| RNA | |||||
|---|---|---|---|---|---|
| Coral ID | Extraction ID | Extraction type | Trial 1 | Trial 2 | Avg (ng/uL) |
| 1431 | 1 | soft | 170 | 170 | 170 |
| 1431 | 1 | hard | 106 | 106 | 106 |
| 1431 | 2 | soft | 162 | 162 | 162 |
| 1431 | 2 | hard | 67.2 | 67.2 | 67.2 |
| 1431 | 3 | soft | 195 | 195 | 195 |
| 1431 | 3 | hard | 116 | 116 | 116 |
| 1431 | 4 | soft | 197 | 197 | 197 |
| 1431 | 4 | hard | 92.4 | 92.4 | 92.4 |
| 1591 | 5 | soft | 32.6 | 32.6 | 32.6 |
| 1591 | 5 | hard | 31.6 | 31.6 | 31.6 |
| 1591 | 6 | soft | 36 | 36.2 | 36.1 |
| 1591 | 6 | hard | 54.2 | 54.2 | 54.2 |
| 1591 | 7 | soft | 83.8 | 83.8 | 83.8 |
| 1591 | 7 | hard | 53.8 | 53.8 | 53.8 |
| 1591 | 8 | soft | 102 | 102 | 102 |
| 1591 | 8 | hard | 75.8 | 75.8 | 75.8 |
General notes:
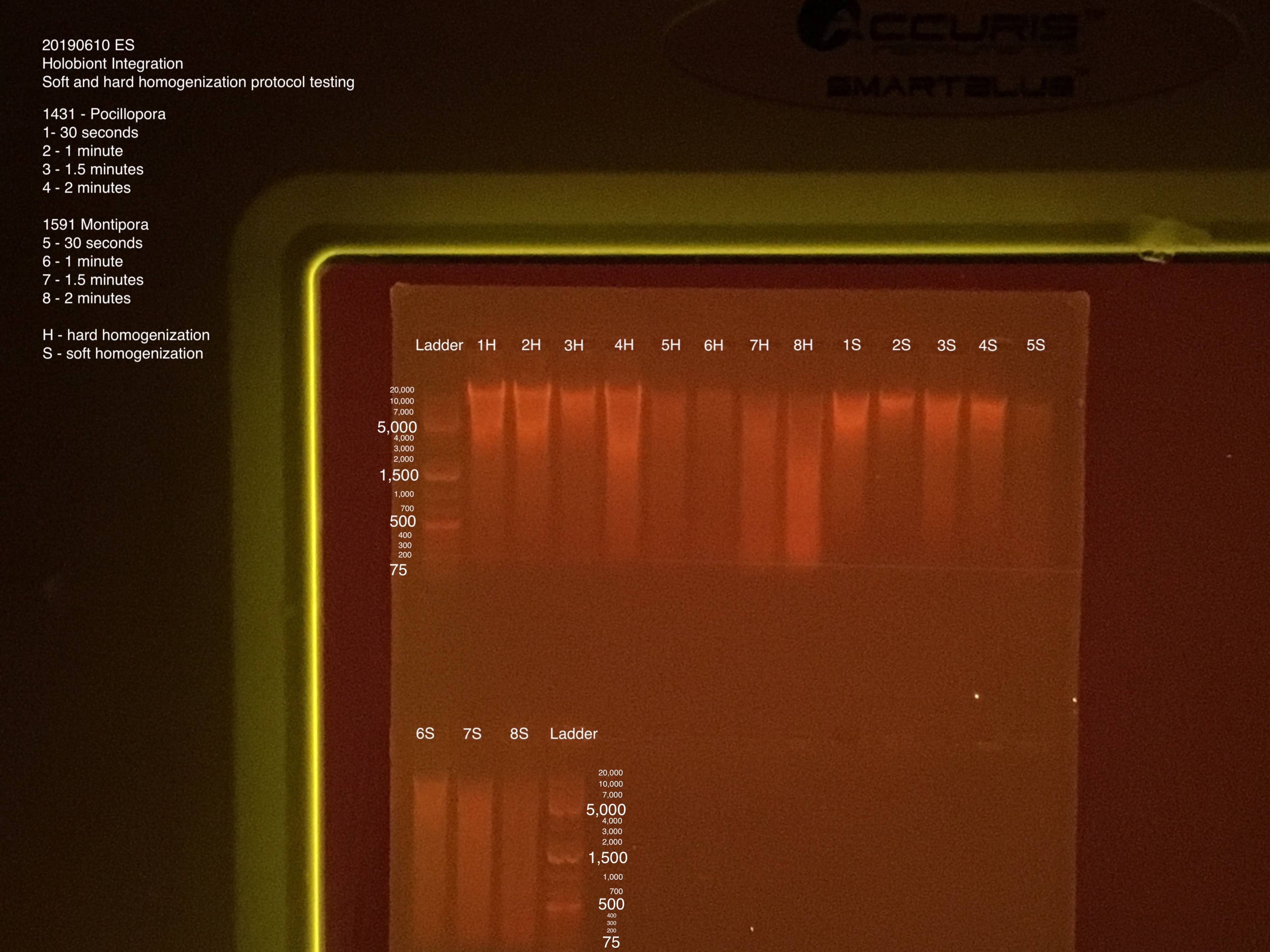
- Used a 2nd comb to fit all samples into gel
- Much easier to keep track of samples when labeled 1-8 S/H instead of using fragment ID; only the final tubes have the all the information on them
- Gel harden start at 3:20; qubit end at 4:30
- Gel start 5:20, end 6:05; run at 100V for 45 minutes
Gel Electrophoresis (Protocol):
- 100V for 45 minutes
- 75 uL TAE buffer + 0.75 g + 5 uL of Biotium gel RED gel stain to make a 1.5% gel
- 5 uL samples + 1 uL 6x purple loading dye
- 4 uL GeneRuler 1 kb Plus DNA ladder + 1 uL 6x purple loading dye
Gel order: Ladder, 1-8H, 1-8S, Ladder
TapeStation (Protocol)
- 2 screentapes used (each tape has 16 slots)
- 1st round: Ladder, 1-6H; start 5:31 end 5:41
- 2nd round: Ladder, 1-7S, 7H, 8S, 8H; screentape switched halfway through- start 5:58 end 6:08 start 6:09 end 6:15
- Ladder: Agilent RNA screentape ladder
| Extraction # | Coral ID | Species | Extraction Type | RIN^e |
|---|---|---|---|---|
| 1 | 1431 | P. acuta | Soft | 5.4 |
| 1 | 1431 | P. acuta | Hard | 3.0 |
| 2 | 1431 | P. acuta | Soft | 4.3 |
| 2 | 1431 | P. acuta | Hard | 3.4 |
| 3 | 1431 | P. acuta | Soft | 4.3 |
| 3 | 1431 | P. acuta | Hard | 3.2 |
| 4 | 1431 | P. acuta | Soft | 3.6 |
| 4 | 1431 | P. acuta | Hard | 3.5 |
| 5 | 1591 | M. capitata | Soft | 6.6 |
| 5 | 1591 | M. capitata | Hard | 6.3 |
| 6 | 1591 | M. capitata | Soft | 6.9 |
| 6 | 1591 | M. capitata | Hard | 5.7 |
| 7 | 1591 | M. capitata | Soft | 6.6 |
| 7 | 1591 | M. capitata | Hard | 5.4 |
| 8 | 1591 | M. capitata | Soft | 5.9 |
| 8 | 1591 | M. capitata | Hard | 5.0 |
Full report here: Round #1; Round #2
Concluding thoughts:
- Extraction #5S aliquot might have had issues because this sample is faded in the Gel and the TapeStation came back with a low concentration error. This could also be because of the mucus causing issues during extractions. Qubit values were lower but not that low (DNA 50.6; RNA 32.6).
- P. acuta DNA looks best in 2S with quantity and quality. This was 1 minute in soft homogenization step. But, the skeleton looked paler in #2 than #3, which was 1.5 minutes. This is why I think position and size factors in here too.
- M. capitata too mucus-y in soft homogenizations equal to and under 1 minute. Needs at least 1.5 minutes to break down tissue.
- M. capitata DNA faded in gel for both soft and hard, but M. capitata had higher quality RNA than P. acuta.
- Hard homogenization step was done with a vortex because of equipment issues (see bullet points at beginning of post) so this could change later.
