KBay Bleaching Pairs 16S Processing
KBay Bleaching Pairs 16S Processing
Processing with Ariana and Kevin’s samples. Spreadsheet for sample names and plate maps found here.
Putnam Lab 16S protocol found here.
Trial run
Master mix calculations:
| (ul) | # of rxns | (ul) to add | |
|---|---|---|---|
| Phusion MM | 12.5 | 43.5 | 543.75 |
| 806RB primer | 0.5 | 43.5 | 21.75 |
| 515F primer | 0.5 | 43.5 | 21.75 |
| Ultra pure H2O | 10.5 | 43.5 | 456.75 |
Platemap (plate 1)
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | KW-10 | KW-10 | KW-10 | KW-11 | KW-11 | KW-11 | KW-12 | KW-12 | KW-12 | KW-13 | KW-13 | KW-13 |
| B | AH-2 | AH-2 | AH-2 | AH-3 | AH-3 | AH-3 | AH-4 | AH-4 | AH-4 | AH-5 | AH-5 | AH-5 |
| C | ES-2 | ES-2 | ES-2 | ES-4 | ES-4 | ES-4 | ES-6 | ES-6 | ES-6 | ES-16 | ES-16 | ES-16 |
| D | POS(+) | POS(+) | POS(+) | NEG(-) | NEG(-) | NEG(-) |
Positive control: M. capitata sample from Holobiont Integration M2410 from well B1 from dilution plate 2.
Negative control: Ultrapure water
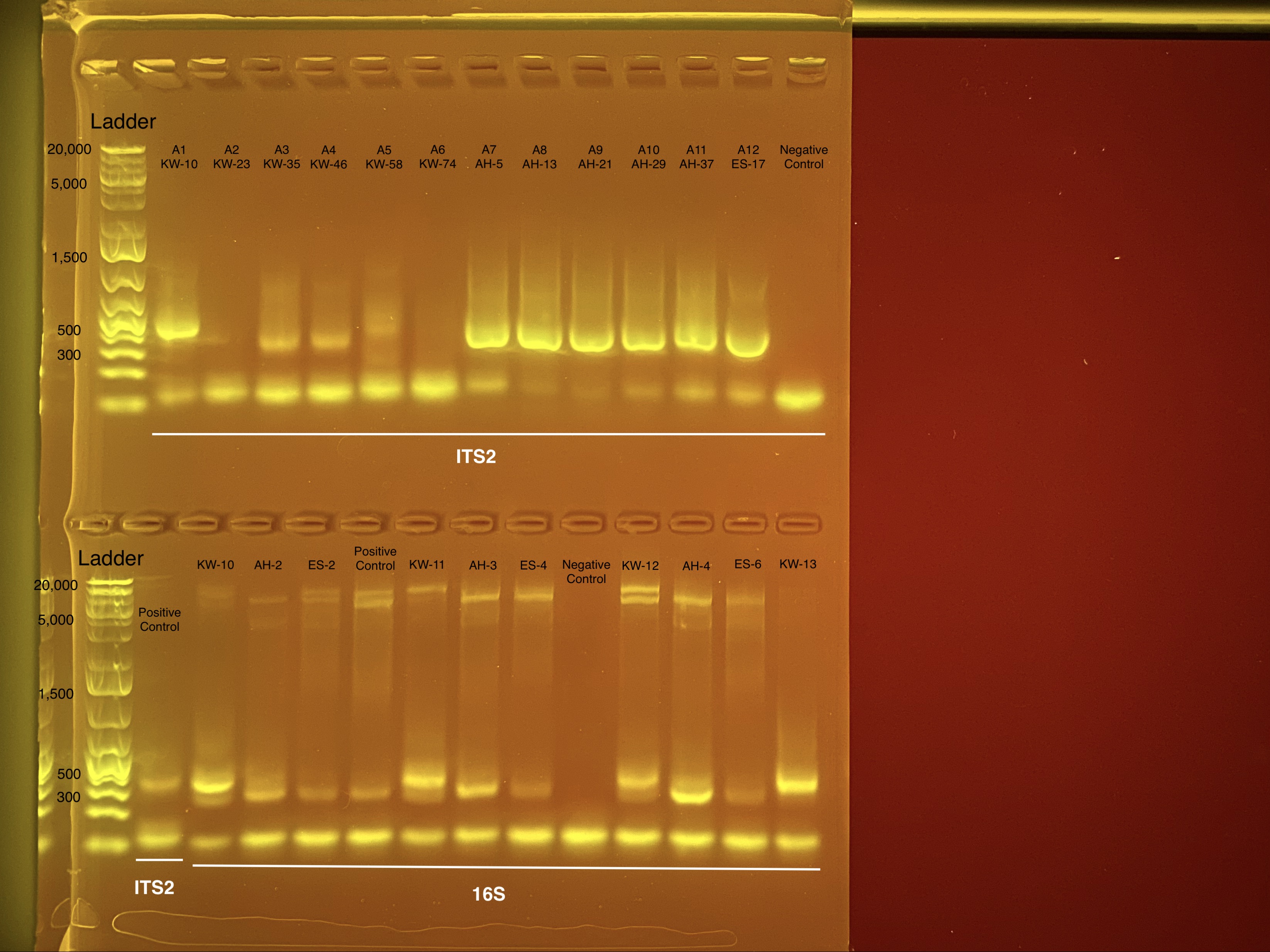
Initially we were worried about the difference in band size between KW and AH/ES samples (KW appears to be closer to 400 bp and AH/ES bands seem to be closer to 300 bp long). This could be because AH and ES samples are picking up more of the 18S and mitochondria regions than the 16S regions. However, the positive control was a sample that we have sequenced previously and in the gel image above it looks like the M. capitata ES samples.
Positive control M2410 sequencing details
M2410 = HPW060 (Sequencing ID with URI GSC)
Denoising stats: M2410 was the 57th lowest # of reads after filtering out non-chimeric out of 262 samples.
| Plug ID | Sequencing ID | Input | Filtered | % of input passed filtered | Denoised | Merged | % of input merged | Non-chimeric | % of input non-chimeric |
|---|---|---|---|---|---|---|---|---|---|
| M2410 | HPW060 | 31027 | 22804 | 73.5 | 22472 | 20306 | 65.45 | 19673 | 63.41 |
| Average of all samples | 40776.97 | 29593.68 | 72.49211 | 29411.86 | 27954.59 | 67.92323 | 27939.43 | 67.88873 |
Next steps could be to try a short run with more of the samples from Holobiont Integration - because I only did one of my pervious ones, I don’t know if the faint band is truly reflective of the below average denoising statistics above. We decided to move forward with the protocol as is.
Bacteria / Total:
- M2410: 64.11% of reads are bacteria (12,612 out of 19,673 reads)
- Average sample: 56.12% reads are bacteria
Chloroplast / Total:
- M2410: 340 reads are chloroplast (1.73%)
- Average sample: 2.62% reads are chloroplast
Plate 2, 3
Plate 2 (20211111)
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | KW-10 | KW-10 | KW-10 | 23 | 23 | 23 | 35 | 35 | 35 | 46 | 46 | 46 |
| B | 11 | 11 | 11 | 24 | 24 | 24 | 36 | 36 | 36 | 49 | 49 | 49 |
| C | 12 | 12 | 12 | 29 | 29 | 29 | 37 | 37 | 37 | 51 | 51 | 51 |
| D | 13 | 13 | 13 | 30 | 30 | 30 | 39 | 39 | 39 | 53 | 53 | 53 |
| E | 14 | 14 | 14 | 31 | 31 | 31 | 40 | 40 | 40 | 54 | 54 | 54 |
| F | 15 | 15 | 15 | 32 | 32 | 32 | 41 | 41 | 41 | 55 | 55 | 55 |
| G | 21 | 21 | 21 | 33 | 33 | 33 | 43 | 43 | 43 | 56 | 56 | 56 |
| H | 22 | 22 | 22 | 34 | 34 | 34 | 44 | 44 | 44 | 57 | 57 | 57 |
Plate 3 (20211111)
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 58 | 58 | 58 | 74 | 74 | 74 | 5 | 5 | 5 | 13 | 13 | 13 |
| B | 62 | 62 | 62 | 80 | 80 | 80 | 6 | 6 | 6 | 14 | 14 | 14 |
| C | 63 | 63 | 63 | 87 | 87 | 87 | 7 | 7 | 7 | 16 | 16 | 16 |
| D | 65 | 65 | 65 | 88 | 88 | 88 | 8 | 8 | 8 | 15 | 15 | 15 |
| E | 67 | 67 | 67 | 89 | 89 | 89 | 9 | 9 | 9 | 17 | 17 | 17 |
| F | 68 | 68 | 68 | AH-2 | AH-2 | AH-2 | 10 | 10 | 10 | 18 | 18 | 18 |
| G | 71 | 71 | 71 | 3 | 3 | 3 | 11 | 11 | 11 | 19 | 19 | 19 |
| H | 73 | 73 | 73 | 4 | 4 | 4 | 12 | 12 | 12 | 20 | 20 | 20 |
Strip tubes for positive and negative control
| POS(+) 1 | POS(+) 1 | POS(+) 1 | POS(+) 2 | POS(+) 2 | POS(+) 2 | POS(+) 3 | POS(+) 3 |
|---|---|---|---|---|---|---|---|
| POS(+) 3 | NEG(-) | NEG(-) | NEG(-) |
Master mix calculations:
| (ul) | # of rxns | (ul) to add | |
|---|---|---|---|
| Phusion MM | 12.5 | 215 | 2687.5 |
| 806RB primer | 0.5 | 215 | 107.5 |
| 515F primer | 0.5 | 215 | 107.5 |
| Ultra pure H2O | 10.5 | 215 | 2257.5 |
Positive control details:
| POS(+) 1 | M1095 | Dilution plate 2 well D1 |
|---|---|---|
| POS(+) 2 | M1694 | Dilution plate 2 well E1 |
| POS(+) 3 | M2986 | Dilution plate 2 well F1 |
Plates were left at 4C instead of -20C over the weekend. The gel looks OK, I won’t redo these for now.
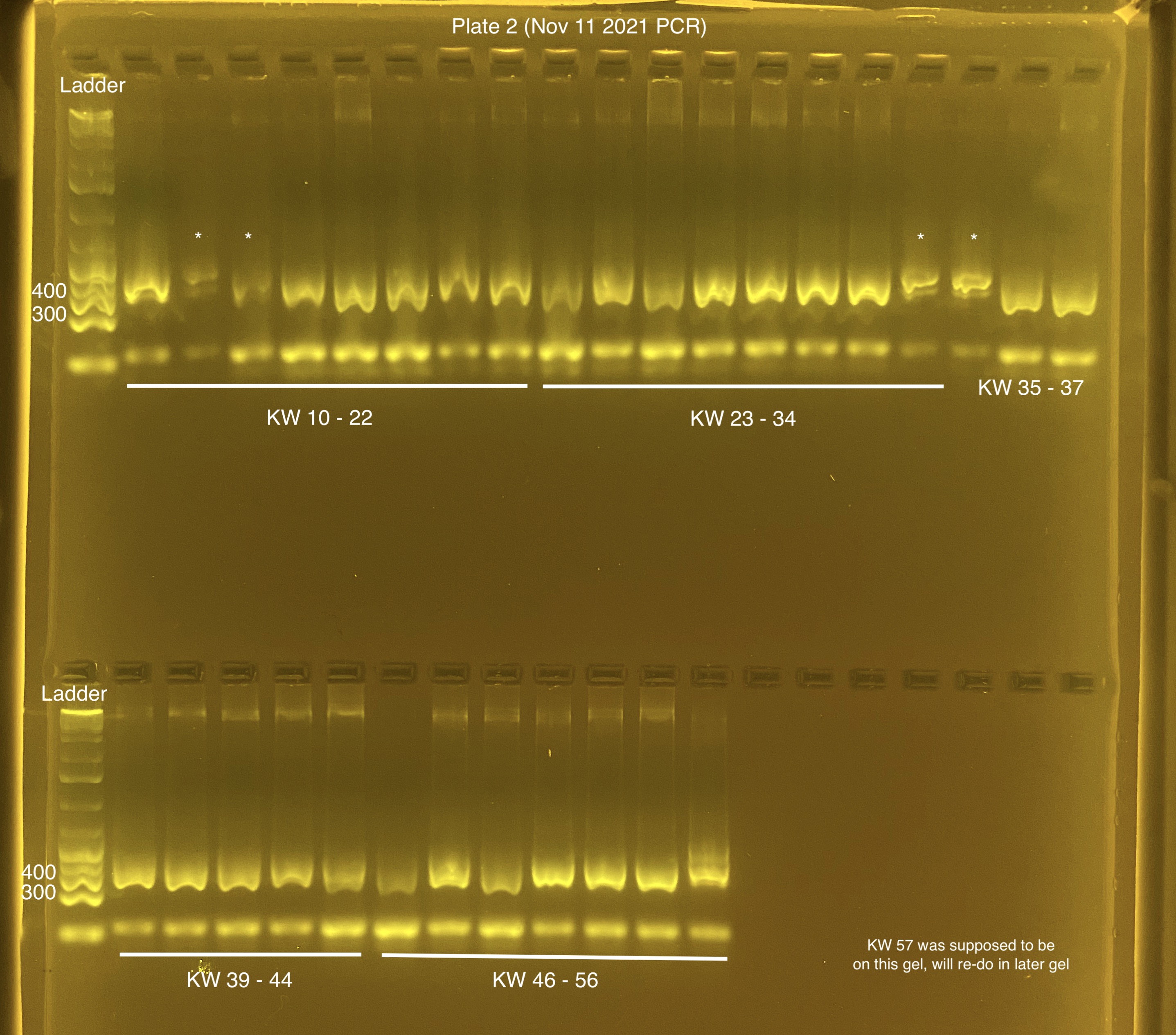
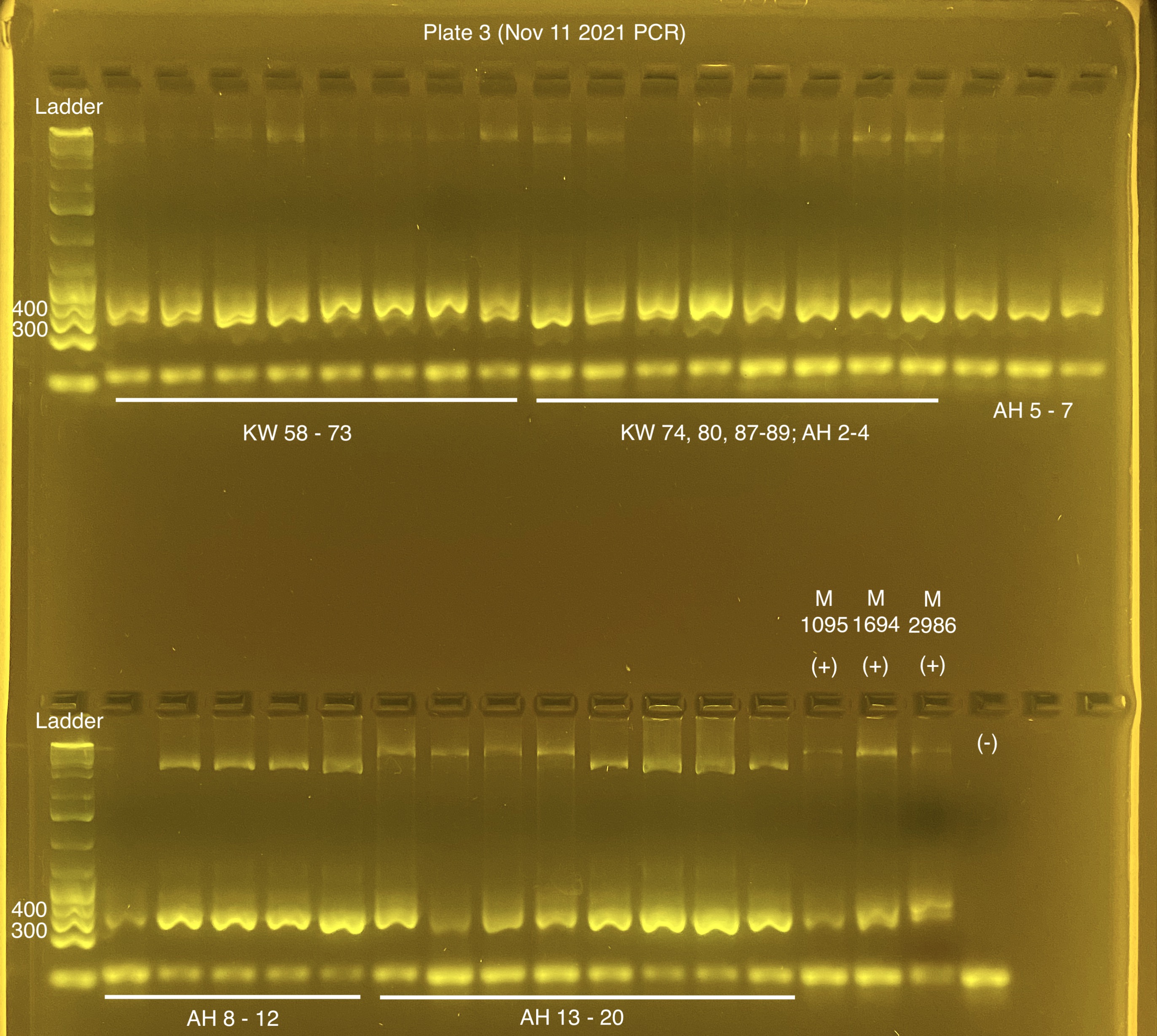
Plate 4, 5
Plate 4 (20211117)
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 21 | 21 | 21 | 29 | 29 | 29 | 37 | 37 | 37 | 17 | 17 | 17 |
| B | 22 | 22 | 22 | 30 | 30 | 30 | 38 | 38 | 38 | 18 | 18 | 18 |
| C | 23 | 23 | 23 | 31 | 31 | 31 | 39 | 39 | 39 | 21 | 21 | 21 |
| D | 24 | 24 | 24 | 32 | 32 | 32 | 40 | 40 | 40 | 22 | 22 | 22 |
| E | 25 | 25 | 25 | 33 | 33 | 33 | ES-2 | ES-2 | ES-2 | 23 | 23 | 23 |
| F | 26 | 26 | 26 | 34 | 34 | 34 | 4 | 4 | 4 | 24 | 24 | 24 |
| G | 27 | 27 | 27 | 35 | 35 | 35 | 6 | 6 | 6 | 25 | 25 | 25 |
| H | 28 | 28 | 28 | 36 | 36 | 36 | 16 | 16 | 16 | 26 | 26 | 26 |
Plate 5 (20211117)
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 28 | 28 | 28 | 37 | 37 | 37 | 45 | 45 | 45 | 55 | 55 | 55 |
| B | 29 | 29 | 29 | 38 | 38 | 38 | 46 | 46 | 46 | 56 | 56 | 56 |
| C | 30 | 30 | 30 | 39 | 39 | 39 | 47 | 47 | 47 | 57 | 57 | 57 |
| D | 31 | 31 | 31 | 40 | 40 | 40 | 50 | 50 | 50 | 59 | 59 | 59 |
| E | 32 | 32 | 32 | 41 | 41 | 41 | 51 | 51 | 51 | NEG(-) | NEG(-) | NEG(-) |
| F | 33 | 33 | 33 | 42 | 42 | 42 | 52 | 52 | 52 | |||
| G | 34 | 34 | 34 | 43 | 43 | 43 | 53 | 53 | 53 | |||
| H | 35 | 35 | 35 | 44 | 44 | 44 | 54 | 54 | 54 |
No positive controls because these were in the first 2 plates already.
Master mix calculations:
| (ul) | # of rxns | (ul) to add | |
|---|---|---|---|
| Phusion MM | 12.5 | 193 | 2412.5 |
| 806RB primer | 0.5 | 193 | 96.5 |
| 515F primer | 0.5 | 193 | 96.5 |
| Ultra pure H2O | 10.5 | 193 | 2026.5 |
Plates left at 4C overnight and gel’d the next day.
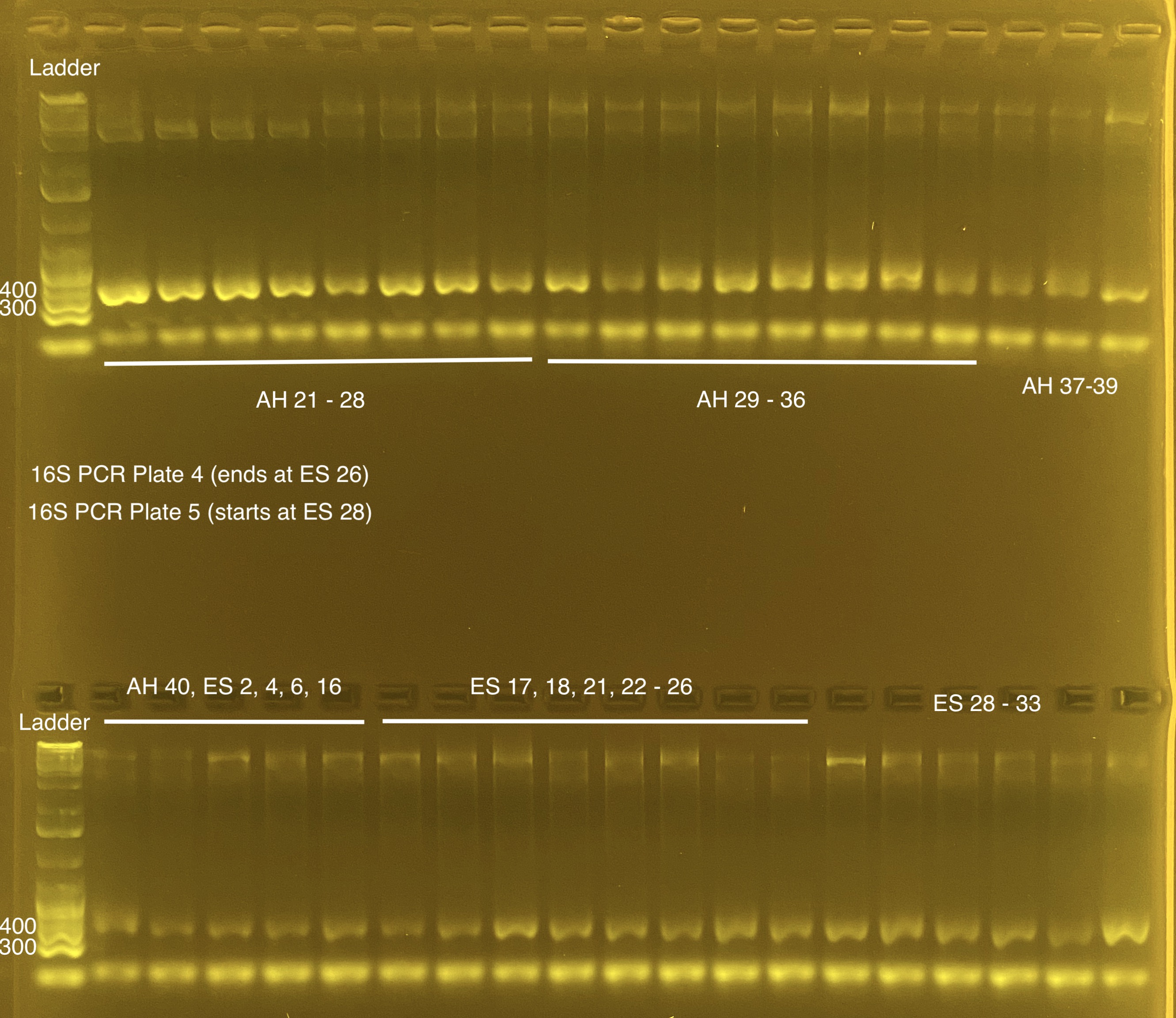
Re-did the gel for some samples from plate 2. These look OK. Ready to submit all of the above samples.
Submit to Janet
Platemaps dropped off to Janet spreadsheet link.
We choose the code WSH (Wong Strand Huffmyer)001 - 096 for the first plate, 101-196 for the second plate, and 201-296 for the third plate.
