HoloInt WGBS Sample Processing
Holobiont Integration WGBS Sample Processing
Test runs were completed prior to sample processing: see notebook post here.
Maggie’s notebook posts for processing Danielle Becker’s samples is here (as another point of reference).
Putnam Lab WGBS Pico Methyl Seq Library Prep Protocol found here.
Dennis’s WGBS test runs (10 pg - 100 ng input) found here.
Zymo Pico Methyl Seq Library Prep Kit link.
Pico Methylation Prep
I’m running my samples with a 10 ng input. I dilute my samples to 5 ng/uL so that I can take 2 uL of DNA and 18 uL Tris HCl for the bisulfite conversion step.
Link to my google spreadsheet for sample processing here (if you don’t already have access, you may request it via a google email address). This includes 5 ng/uL dilution calculations, dilution platemaps, amplification step calculations, indices information, and post-PMS protocol quantification.
For Holobiont Integration, we are processing 5 timepoints: 30 hour, 2 weeks, 4 weeks, 8 weeks, and 12 weeks (n=60).
Notes
- P. acuta 2878 has already been sequenced from this project for the methylation methods comparison paper so we won’t need to do this one again.
- I started with n=3 and n=4 during trial runs but worked my way up to n=8 and n=10 at a time for this protocol. My maximum will probably be 16 samples at a time.
- These samples will be sequenced with K. Wong and my 2nd chapter Hawaii Bleaching Pairs project. This will span across 4 lanes, each project taking up its own lane and HoloInt using 2 lanes.
- HoloInt indices: #1-60
- I give every sample a Pico # which is the number of the pico methylation prep I have done. There are going to be some samples that need to be re-done and for ease of labeling tubes, I will refer to each sample by their Pico # instead of the Plug ID. This will make it easier to set aside the desired tubes for sequencing rather than going by Plug ID and date of PMS protocol.
- We never used index #60 because we sequenced one of the samples the methylation compare project.
- We added M1312 fragment because it is a genome sample that we also want a methylome for.
Post Pico Quantification (Final IDs)
As of 2021-07-20: 59/59 prepped. (The 60th sample has already been sequenced).
See the above linked google spreadsheet for the quantification results of those pico preps that were not final.
2021-03-31 Qubit: DNA BR Standard 1 = 173.28; Standard 2 = 19,833.04
2021-04-06 Qubit: DNA BR Standard 1 = 179.54; Standard 2 = 19,252.29
2021-05-24 Qubit: DNA BR Standard 1 = 194.96; Standard 2 = 21,282.28
2021-06-09 Qubit: DNA BR Standard 1 = 192.07; Standard 2 = 20,942.30
2021-06-10 Qubit: DNA BR Standard 1 = 179.67; Standard 2 = 19,385.82
2021-06-14 Qubit: DNA BR Standard 1 = 190.50; Standard 2 = 20,152.88
2021-07-01 Qubit: DNA BR Standard 1 = 193.88; Standard 2 = 20,559.34
2021-07-13 and 2021-07-14 Qubit: DNA BR Standard 1 = 158.98; Standard 2 = 16,819.25
2021-07-15 Qubit: DNA BR Standard 1 = 185.65; Standard 2 = 19,940.15
2021-07-20 Qubit: DNA BR Standard 1 = 194.00; Standard 2 = 20,712.38
| PLUG_ID | Species | Treatment | Extraction Date | Notes | Soft DNA ng_ul | Final Pico # | Post-Pico Quant (ng/uL) | Indices # | Bp size | Date of Final Pico | Notes |
|---|---|---|---|---|---|---|---|---|---|---|---|
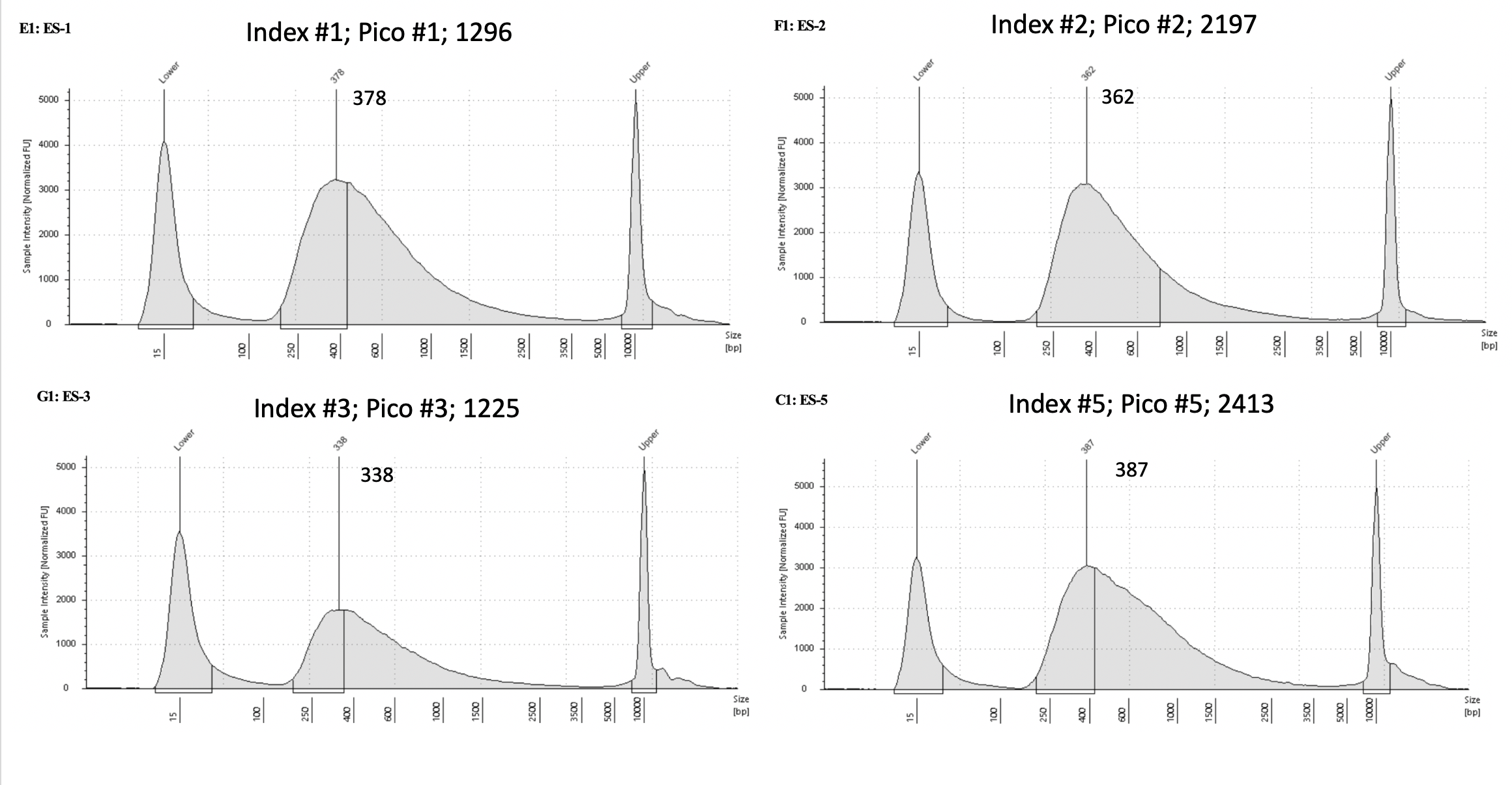
| 1296 | Pacuta | ATHC | 20190729 | 147, 148 - 2nd DNA elution values | 29.6 | 1 | 23.1 | 1 | 378 | 20210331 | |
| 2197 | Pacuta | ATHC | 20190729 | 151, 152 | 40.3 | 2 | 20.4 | 2 | 362 | 20210331 | |
| 1225 | Pacuta | HTAC | 20190730 | 159, 160 | 43.3 | 3 | 13.25 | 3 | 338 | 20210331 | |
| 1312 | Mcapitata | NA | 20210719 | 1312 | 49.8 | 82 | 8.17 | 4 | 324 | 20210720 | M.cap bleached genome fragment |
| 2413 | Pacuta | ATAC | 20190809 | 333, 334 | 59.1 | 5 | 21.2 | 5 | 387 | 20210331 | |
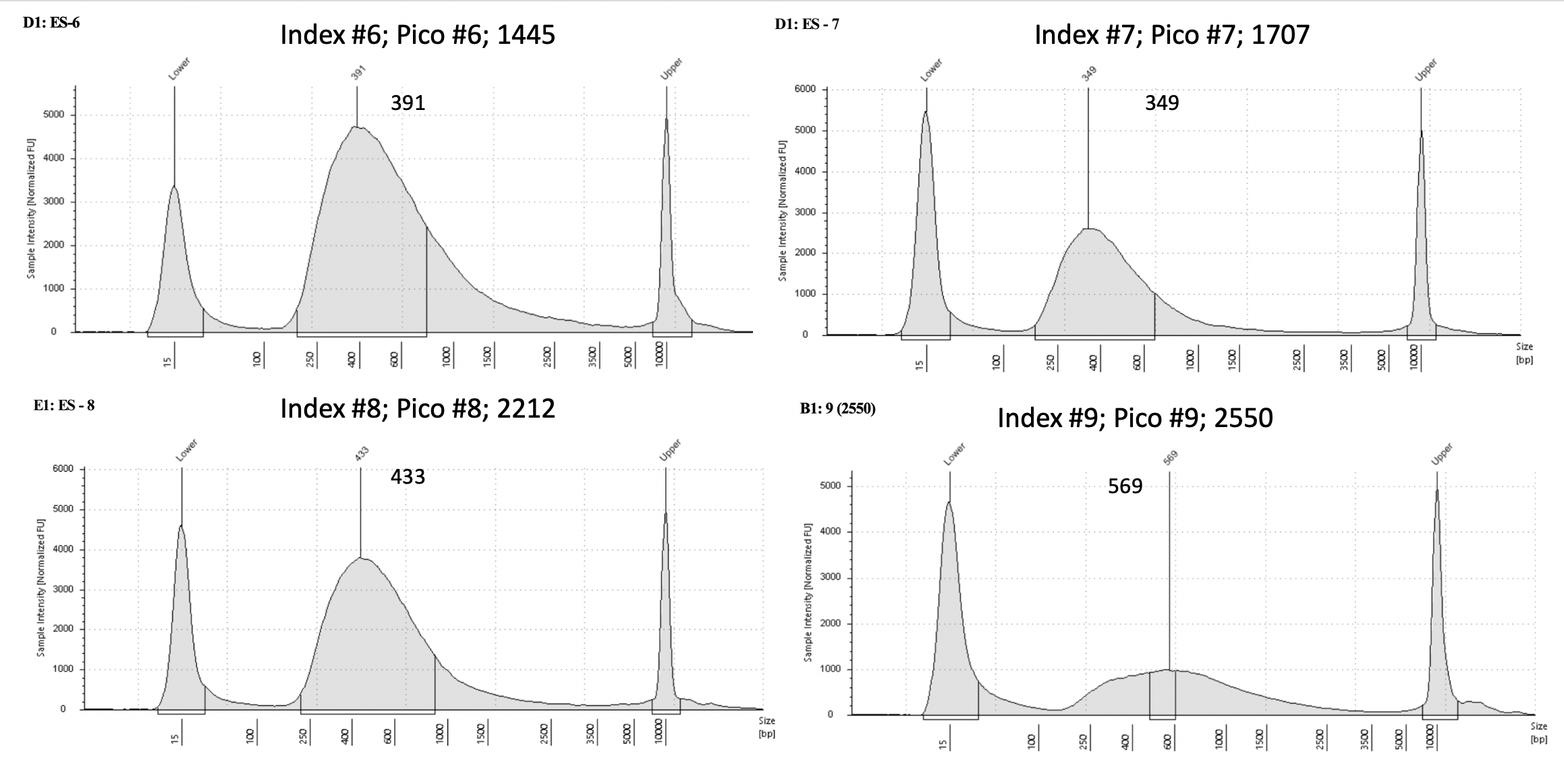
| 1445 | Pacuta | ATAC | 20190826 | 375, 376 | 41.4 | 6 | 16.95 | 6 | 391 | 20210331 | |
| 1707 | Pacuta | HTAC | 20190905 | 445, 446 | 96.6 | 7 | 12.65 | 7 | 349 | 20210331 | |
| 2212 | Pacuta | ATHC | 20190903 | 449, 450 | 34.2 | 8 | 24.9 | 8 | 433 | 20210331 | |
| 2550 | Pacuta | ATHC | 20190724 | 117, 118 | 38.6 | 9 | 7.97 | 9 | 569 | 20210406 | |
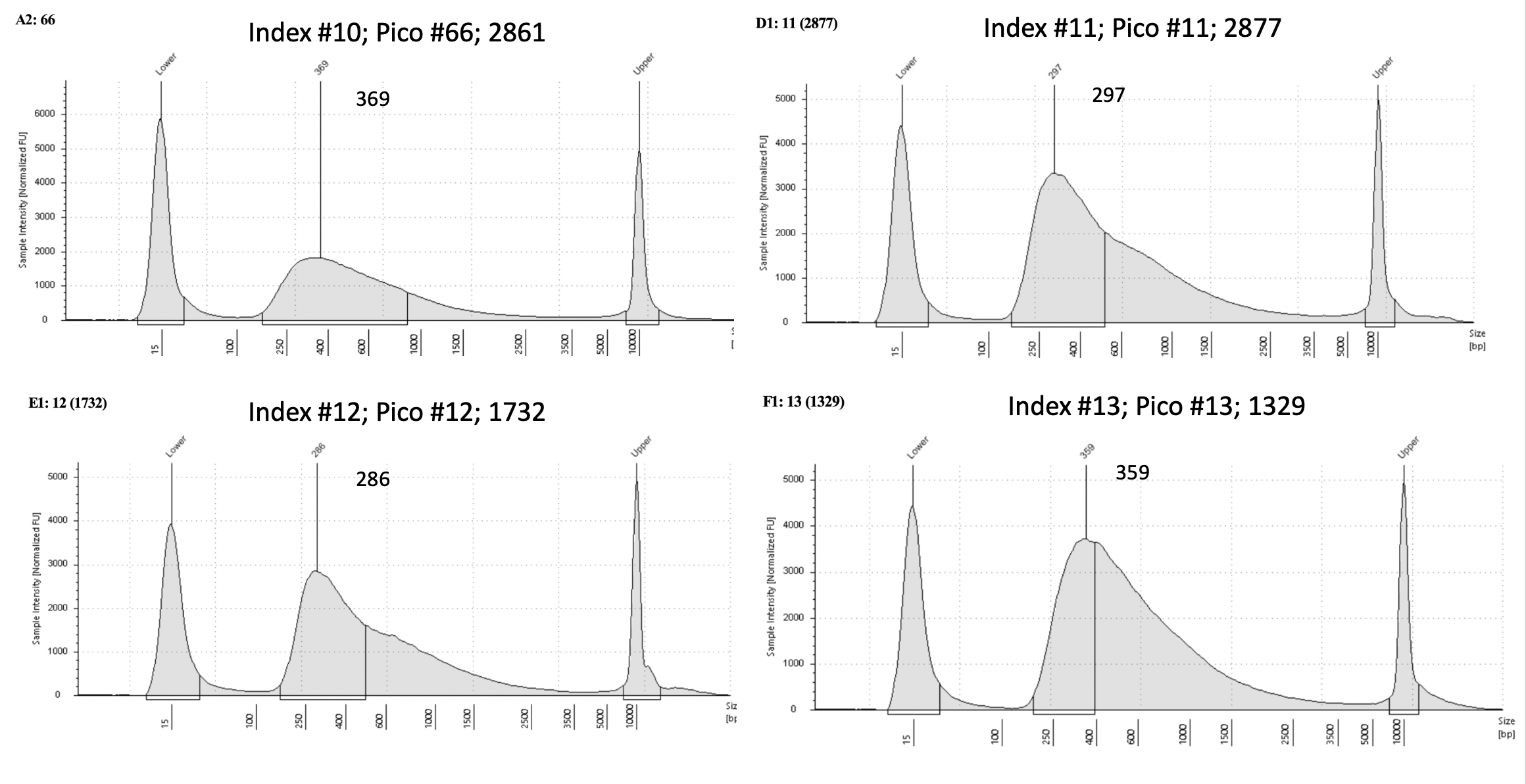
| 2861 | Pacuta | ATHC | 20190730 | 161, 162 | 34.4 | 66 | 15.15 | 10 | 369 | 20210713 | |
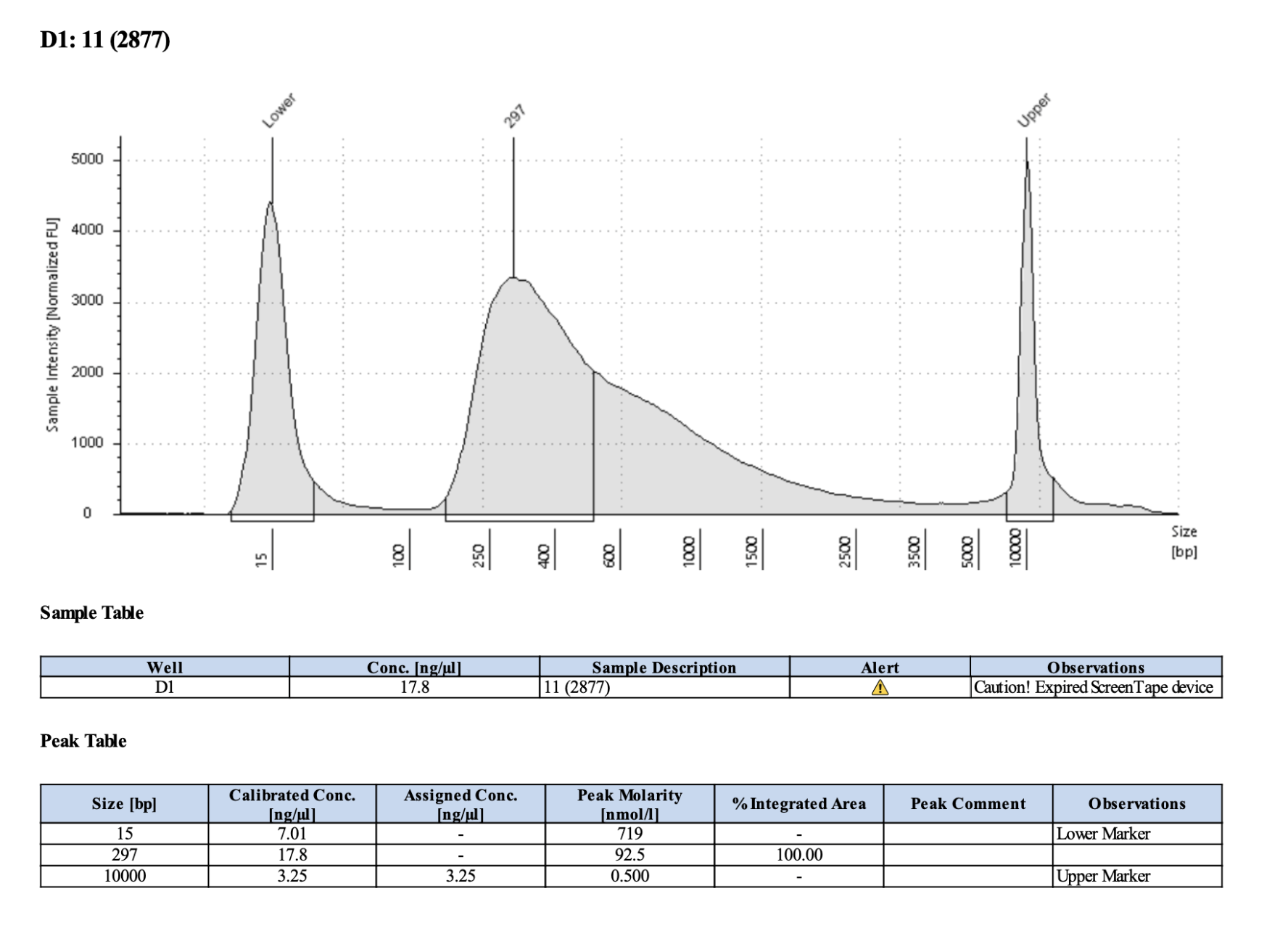
| 2877 | Pacuta | ATHC | 20190731 | 213, 214 | 129.5 | 11 | 22.2 | 11 | 297 | 20210406 | |
| 1732 | Pacuta | HTHC | 20190724 | 111, 112 | 46.2 | 12 | 18.55 | 12 | 286 | 20210406 | |
| 1329 | Pacuta | HTAC | 20191204 | 641, 642 | 41.7 | 13 | 22.7 | 13 | 359 | 20210406 | |
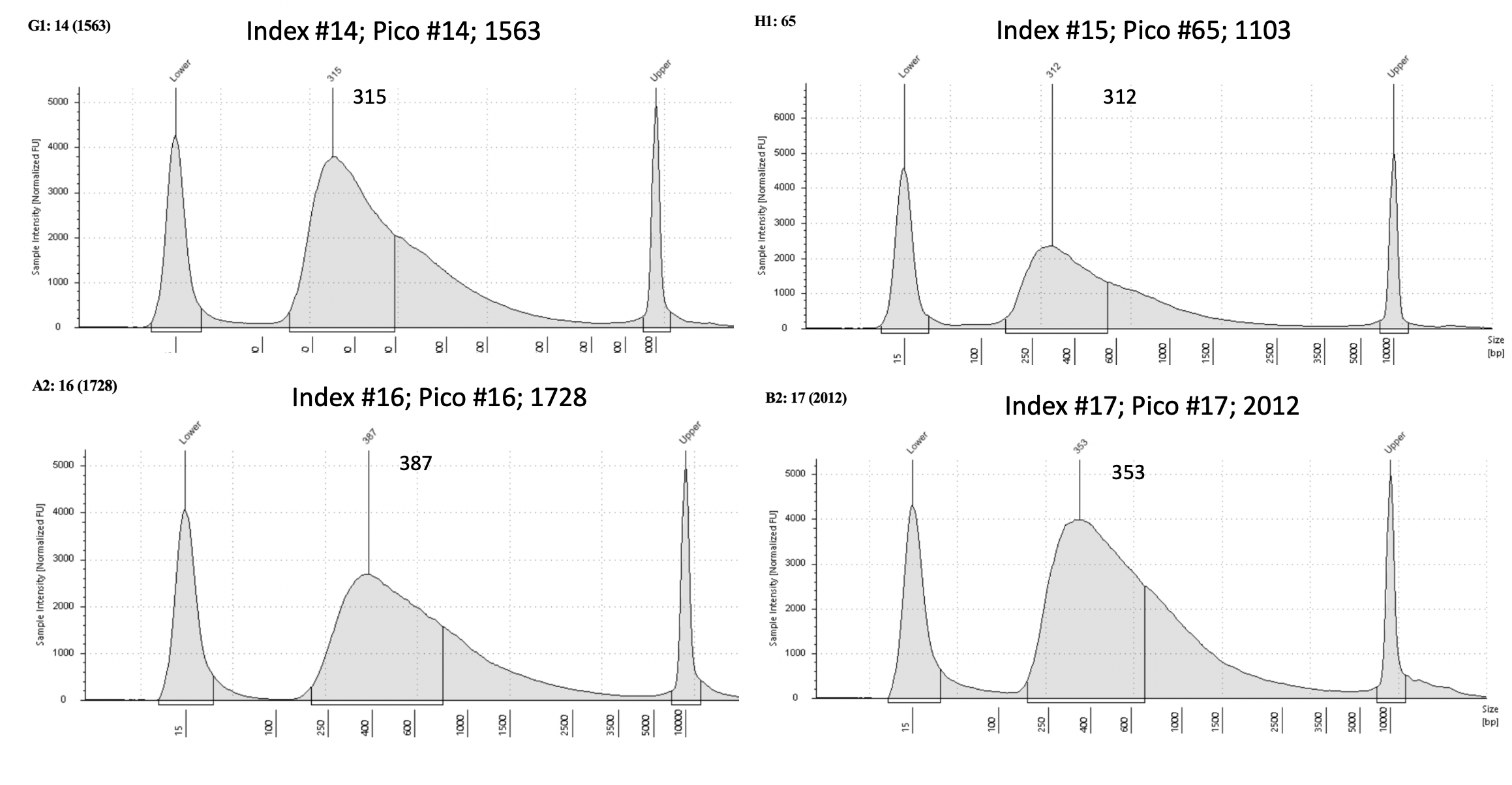
| 1563 | Pacuta | ATAC | 20190905 | 447, 448 | 42.3 | 14 | 24 | 14 | 315 | 20210406 | |
| 1103 | Pacuta | ATAC | 20190718 | 25, 26 | 56.6 | 65 | 18.25 | 15 | 312 | 20210713 | |
| 1728 | Pacuta | HTAC | 20190806 | 293, 294 | 108 | 16 | 19.35 | 16 | 387 | 20210406 | |
| 2012 | Pacuta | ATAC | 20190730 | 203, 204 | 18.9 | 17 | 23.8 | 17 | 353 | 20210406 | |
| 1051 | Pacuta | ATAC | 20190731 | 209, 210 | 86.2 | 18 | 18.3 | 18 | 431 | 20210406 | |
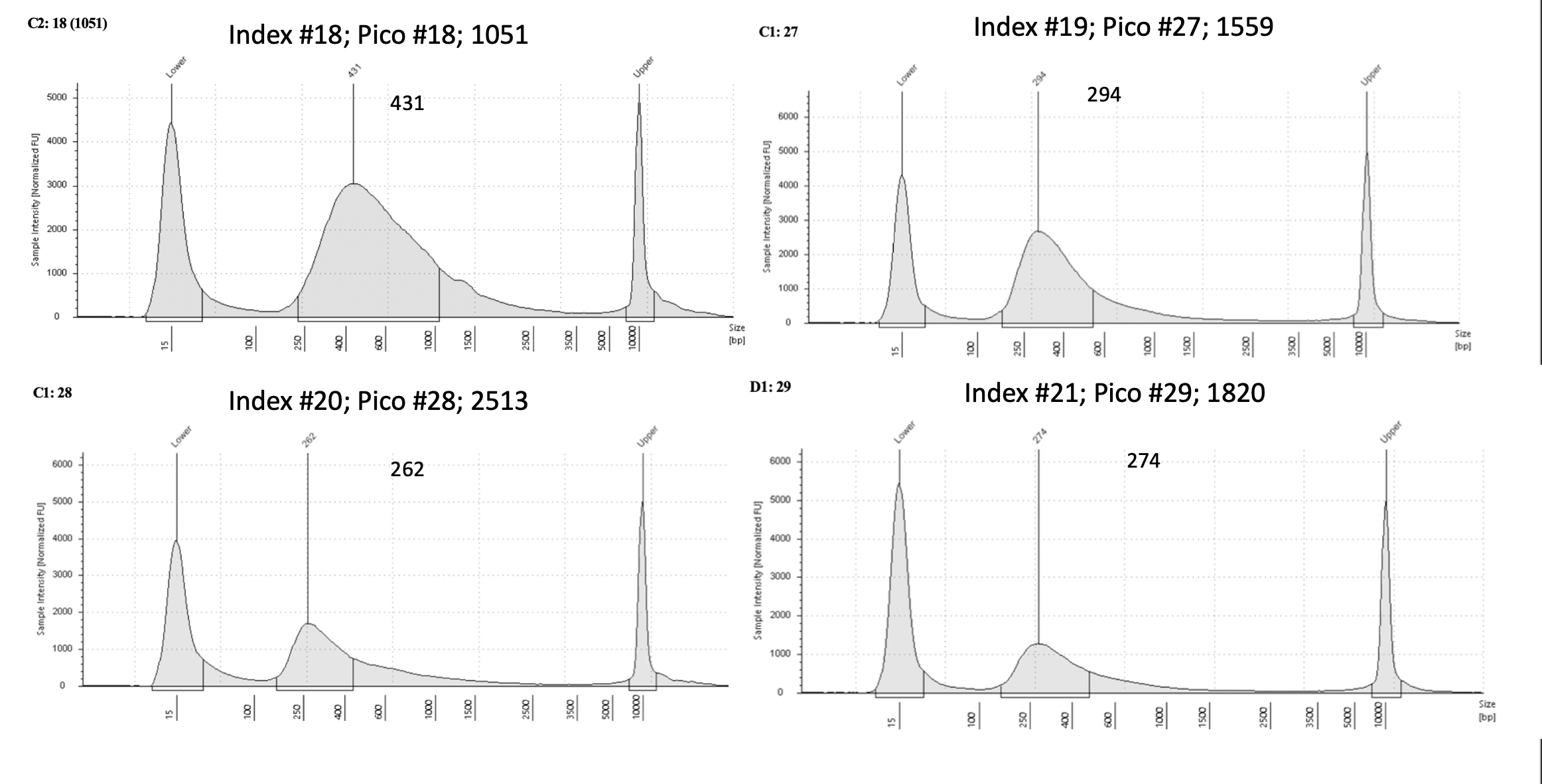
| 1559 | Pacuta | ATAC | 20190725 | 129, 130 | 50 | 27 | 18.05 | 19 | 294 | 20210609 | |
| 2513 | Pacuta | HTAC | 20190722 | 65, 66 | 22.9 | 28 | 9.2 | 20 | 262 | 20210410 | |
| 1820 | Pacuta | HTHC | 20190815 | 353, 354 | 35.4 | 29 | 5.67 | 21 | 274 | 20210410 | |
| 1238 | Pacuta | HTHC | 20190725 | 125, 126 | 43.1 | 30 | 10.2 | 22 | 293 | 20210614 | |
| 2064 | Pacuta | HTAC | 20190730 | 167, 168 | 28.9 | 83 | 23.3 | 23 | 326 | 20210720 | |
| 1168 | Pacuta | HTHC | 20190730 | 171, 172 | 15.95 | 32 | 5.88 | 24 | 274 | 20210410 | |
| 1459 | Pacuta | ATHC | 20190722 | 53, 54 | 57.2 | 68 | 18.6 | 25 | 286 | 20210714 | |
| 1765 | Pacuta | HTAC | 20190725 | 119, 120 | 76 | 69 | 26 | 26 | 306 | 20210714 | |
| 2072 | Pacuta | HTAC | 20190814 | 343, 344 | 41.3 | 70 | 12.6 | 27 | 307 | 20210714 | |
| 2304 | Pacuta | HTHC | 20190722 | 81, 82 | 25.1 | 38 | 9.28 | 28 | 298 | 20210609 | |
| 1184 | Pacuta | HTHC | 20190731 | 183, 184 | 49 | 84 | 21 | 29 | 355 | 20210720 | |
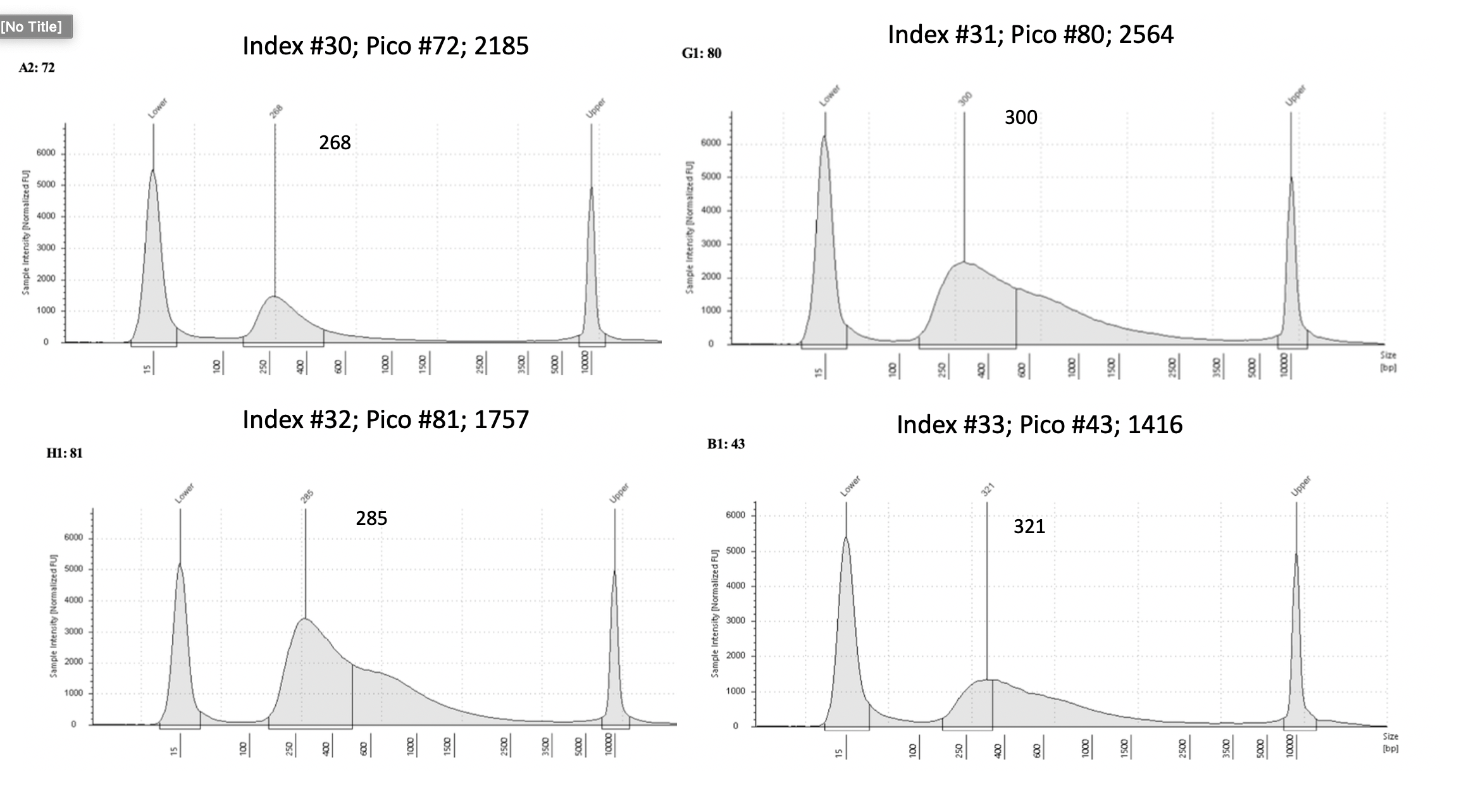
| 2185 | Pacuta | HTHC | 20191009 | 499, 500 | 26.8 | 72 | 5.25 | 30 | 268 | 20210714 | |
| 2564 | Pacuta | ATHC | 20190726 | 139, 140 | 34.1 | 80 | 16.45 | 31 | 300 | 20210715 | |
| 1757 | Pacuta | ATAC | 20190731 | 197, 198 | 48 | 81 | 21.1 | 32 | 285 | 20210715 | |
| 1416 | Pacuta | HTHC | 20191009 | 497, 498 | 16.75 | 43 | 8.57 | 33 | 321 | 20210614 | |
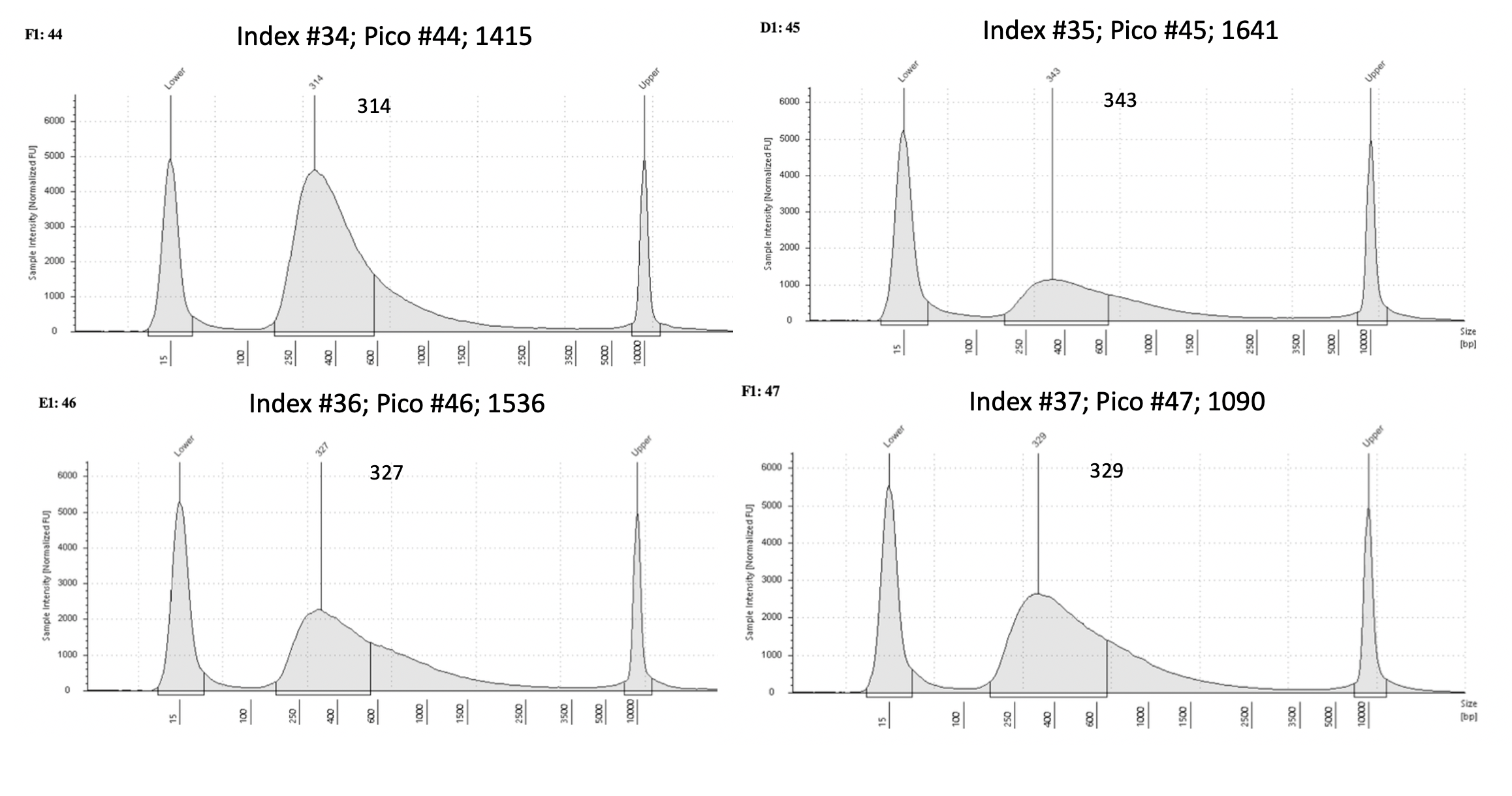
| 1415 | Pacuta | HTHC | 20190815 | 359, 360 | 53.6 | 44 | 19.85 | 34 | 308 | 20210614 | |
| 1641 | Pacuta | ATAC | 20190720 | 39, 40 | 34.5 | 45 | 7.15 | 35 | 343 | 20210614 | |
| 1536 | Pacuta | HTAC | 20190725 | 121, 122 | 79.5 | 46 | 14.65 | 36 | 327 | 20210614 | |
| 1090 | Pacuta | HTHC | 20190807 | 313, 314 | 31.5 | 47 | 15.7 | 37 | 329 | 20210614 | |
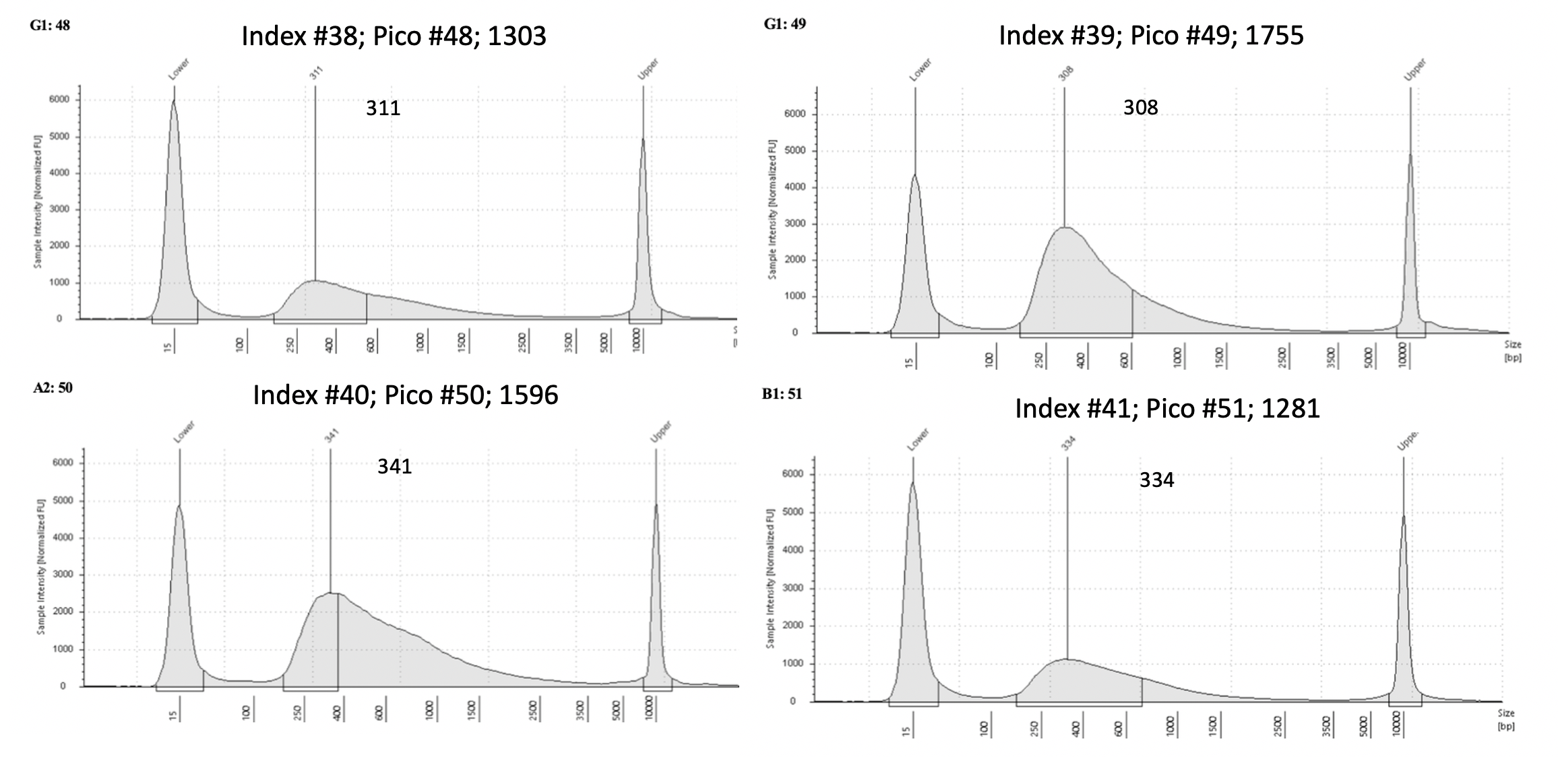
| 1303 | Pacuta | HTAC | 20190726 | 143, 144 | 33.6 | 48 | 6.83 | 38 | 311 | 20210614 | |
| 1755 | Pacuta | ATAC | 20190801 | 251, 252 | 19.25 | 49 | 24.1 | 39 | 314 | 20210614 | |
| 1596 | Pacuta | HTAC | 20191009 | 503, 504 | 27.8 | 50 | 19.1 | 40 | 341 | 20210614 | |
| 1281 | Pacuta | ATHC | 20190924 | 457, 458 | 34.1 | 51 | 7.11 | 41 | 334 | 20210630 | |
| 1571 | Pacuta | HTAC | 20191121 | 581, 582 | 61.5 | 52 | 13.05 | 42 | 316 | 20210630 | |
| 2300 | Pacuta | HTHC | 20190731 | 181, 182 | 62.1 | 53 | 11.5 | 43 | 311 | 20210630 | |
| 1427 | Pacuta | HTHC | 20190807 | 311, 312 | 42.9 | 54 | 8.7 | 44 | 280 | 20210630 | |
| 1205 | Pacuta | ATHC | 20190720 | 45, 46 | 34.6 | 55 | 11.25 | 45 | 312 | 20210630 | |
| 2879 | Pacuta | ATHC | 20190930 | 483, 484 | 34.1 | 56 | 10.65 | 46 | 305 | 20210630 | |
| 1709 | Pacuta | HTHC | 20191121 | 591, 592 | 28.8 | 57 | 13.7 | 47 | 331 | 20210630 | |
| 1777 | Pacuta | ATAC | 20190930 | 481, 482 | 55.9 | 58 | 8.62 | 48 | 309 | 20210630 | |
| 2306 | Pacuta | ATAC | 20190926 | 465, 466 | 73.1 | 59 | 20.6 | 49 | 308 | 20210713 | |
| 1582 | Pacuta | HTAC | 20190926 | 473, 474 | 34.7 | 60 | 28.2 | 50 | 298 | 20210713 | |
| 1647 | Pacuta | HTAC | 20190926 | 469, 470 | 98.1 | 61 | 20 | 51 | 285 | 20210713 | |
| 1159 | Pacuta | ATAC | 20190720 | 37, 38 | 53.1 | 62 | 11.5 | 52 | 308 | 20210713 | |
| 1487 | Pacuta | HTAC | 20190807 | 317, 318 | 37.2 | 63 | 19.5 | 53 | 312 | 20210713 | |
| 2668 | Pacuta | ATHC | 20191001 | 489, 490 | 28.3 | 64 | 20.9 | 54 | 291 | 20210713 | |
| 1047 | Pacuta | ATAC | 20190808 | 325, 326 | 57.5 | 75 | 5.36 | 55 | 336 | 20210715 | |
| 1059 | Pacuta | ATAC | 20190826 | 413, 414 | 27 | 76 | 20 | 56 | 332 | 20210715 | |
| 2409 | Pacuta | ATHC | 20190808 | 323, 324 | 73.5 | 77 | 16.1 | 57 | 334 | 20210715 | |
| 2087 | Pacuta | HTHC | 20190826 | 415, 416 | 50.5 | 78 | 4.72 | 58 | 396 | 20210715 | |
| 1147 | Pacuta | ATHC | 20191206 | 659, 660 | 57.5 | 79 | 21.1 | 59 | 285 | 20210715 | |
| 2878 | Pacuta | ATHC | 20190807 | 319, 320 | 48.1 | NA | NA | NA | NA | NA | Already sequenced in meth compare project |
WGBS processing notes
Pico #19-26 done on 2021-05-24: KAPA hifi mix was used and funky contamination.. see tapestation. These were re-done in Pico #27-34.
Pico #27-42 were all low amplification. Re-amps were done on all except for Pico#40. Re-amps failed except for Pico#28.
Soft DNA E2 tubes missing for: 2409 (Ext ID 323, 324), 1047 (Ext ID 325, 326), 1059 (Ext ID 413, 414), and 2087 (Ext ID 415, 416). So we used the E1 tubes for those samples (qubit below):
- Soft DNA E1 2409 = 531 ng/uL
- Soft DNA E1 1047 = 931 ng/uL
- Soft DNA E1 1059 = 208 ng/uL
- Soft DNA E1 2087 = 524 ng/uL
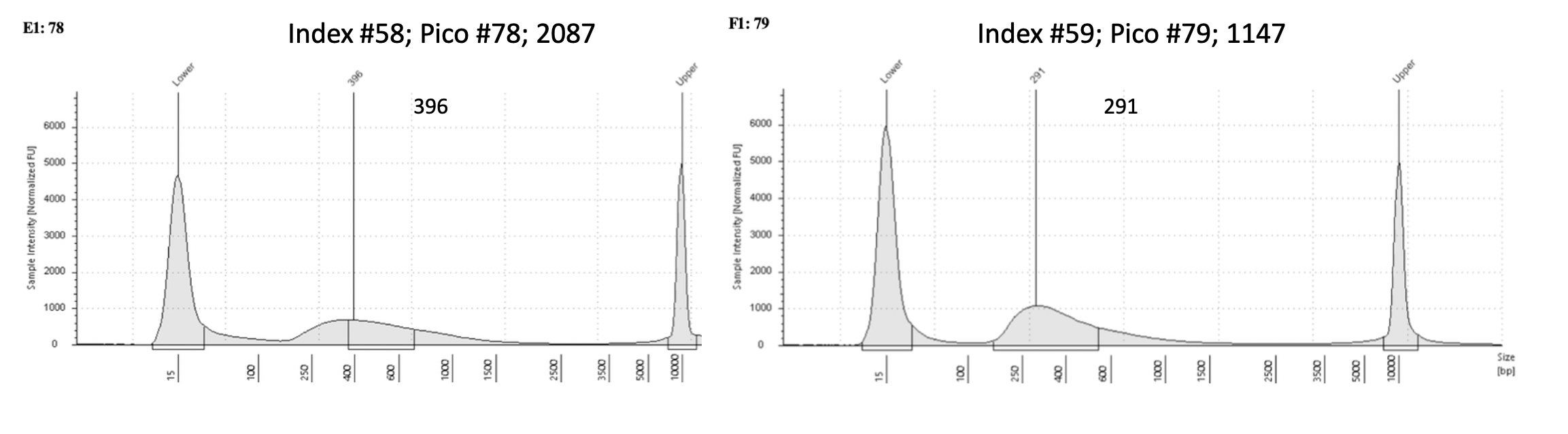
Tapestation examples
Example of acceptable tapestation result:
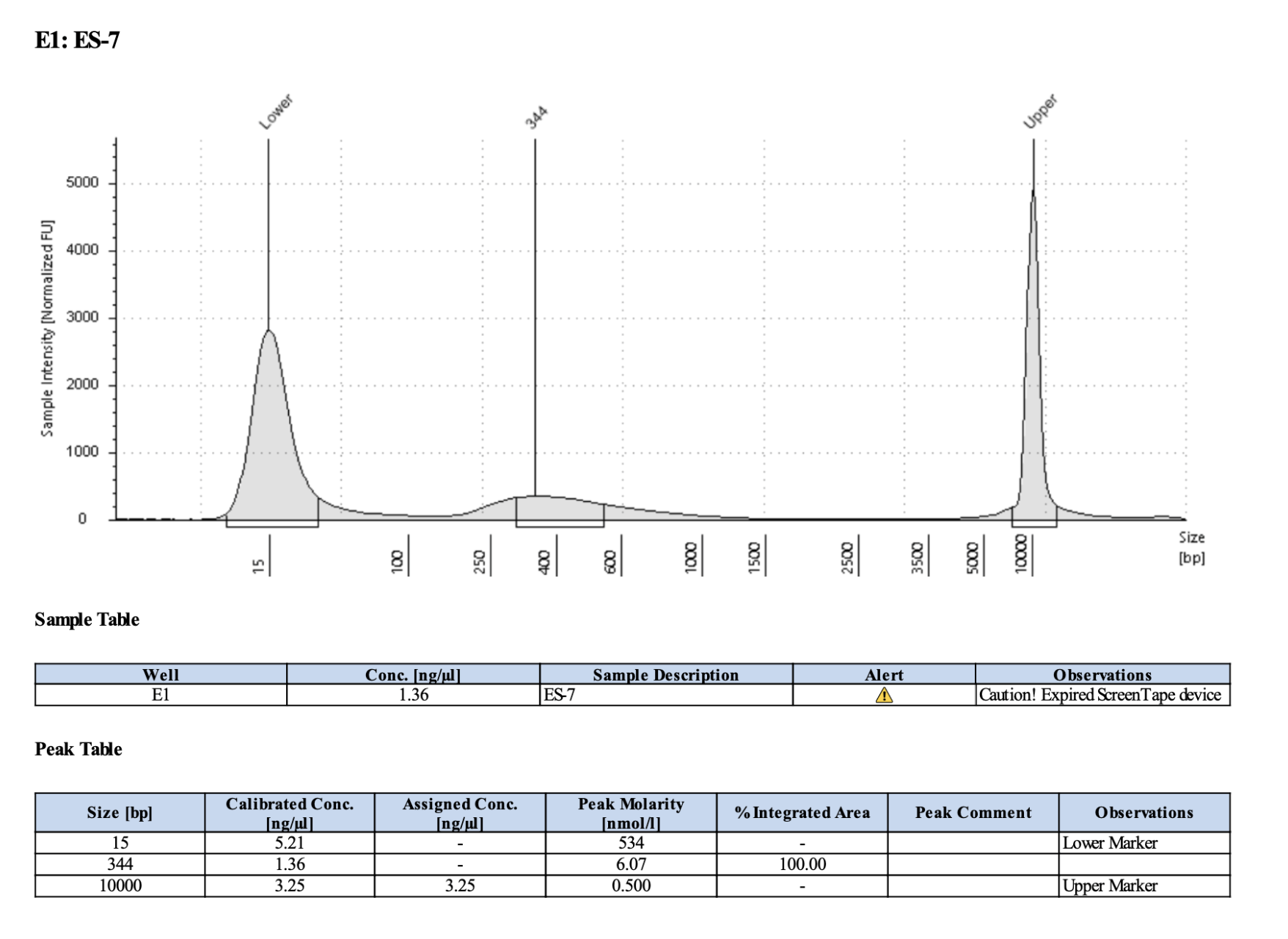
Example of one I would re-do:
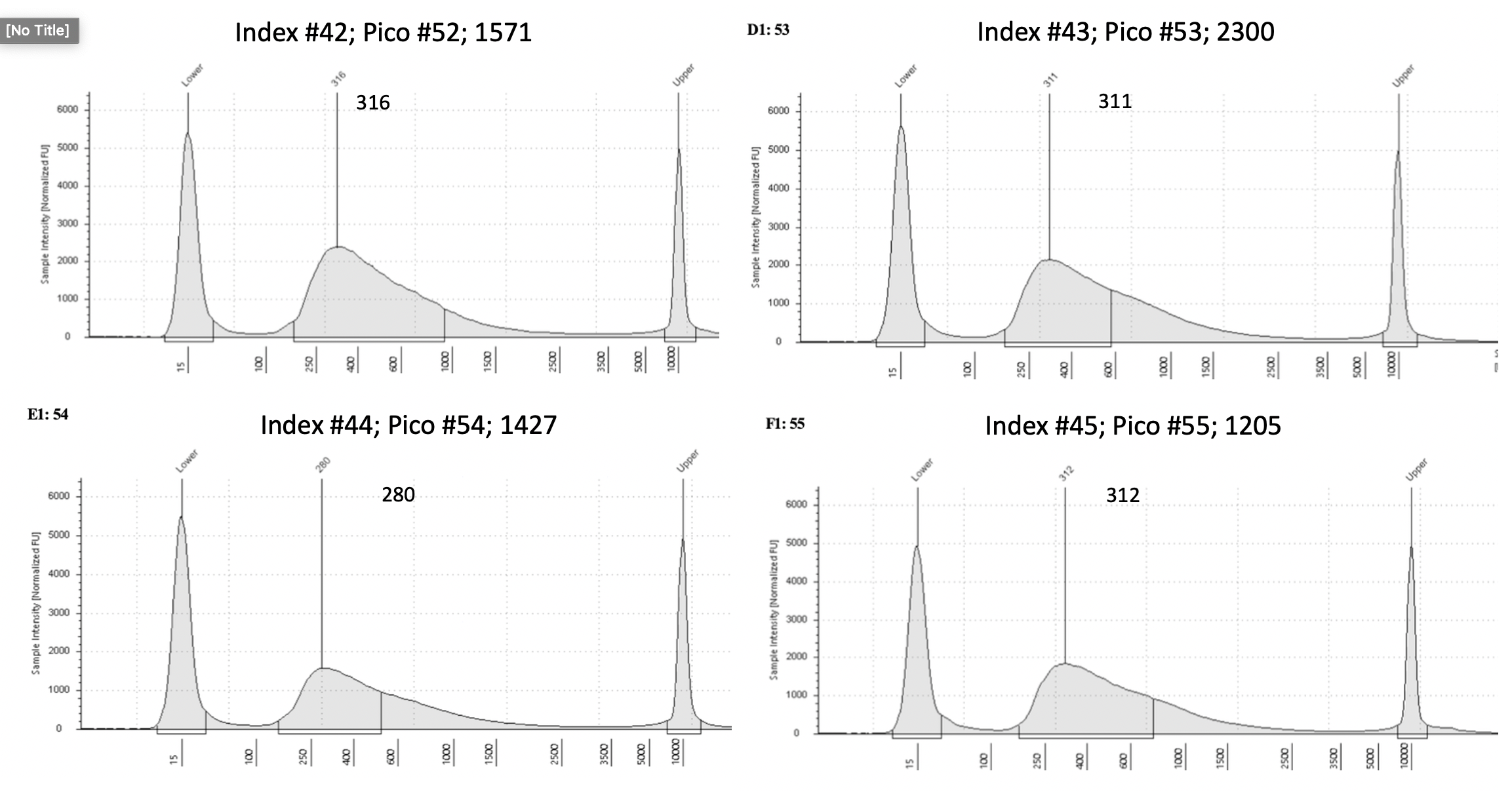
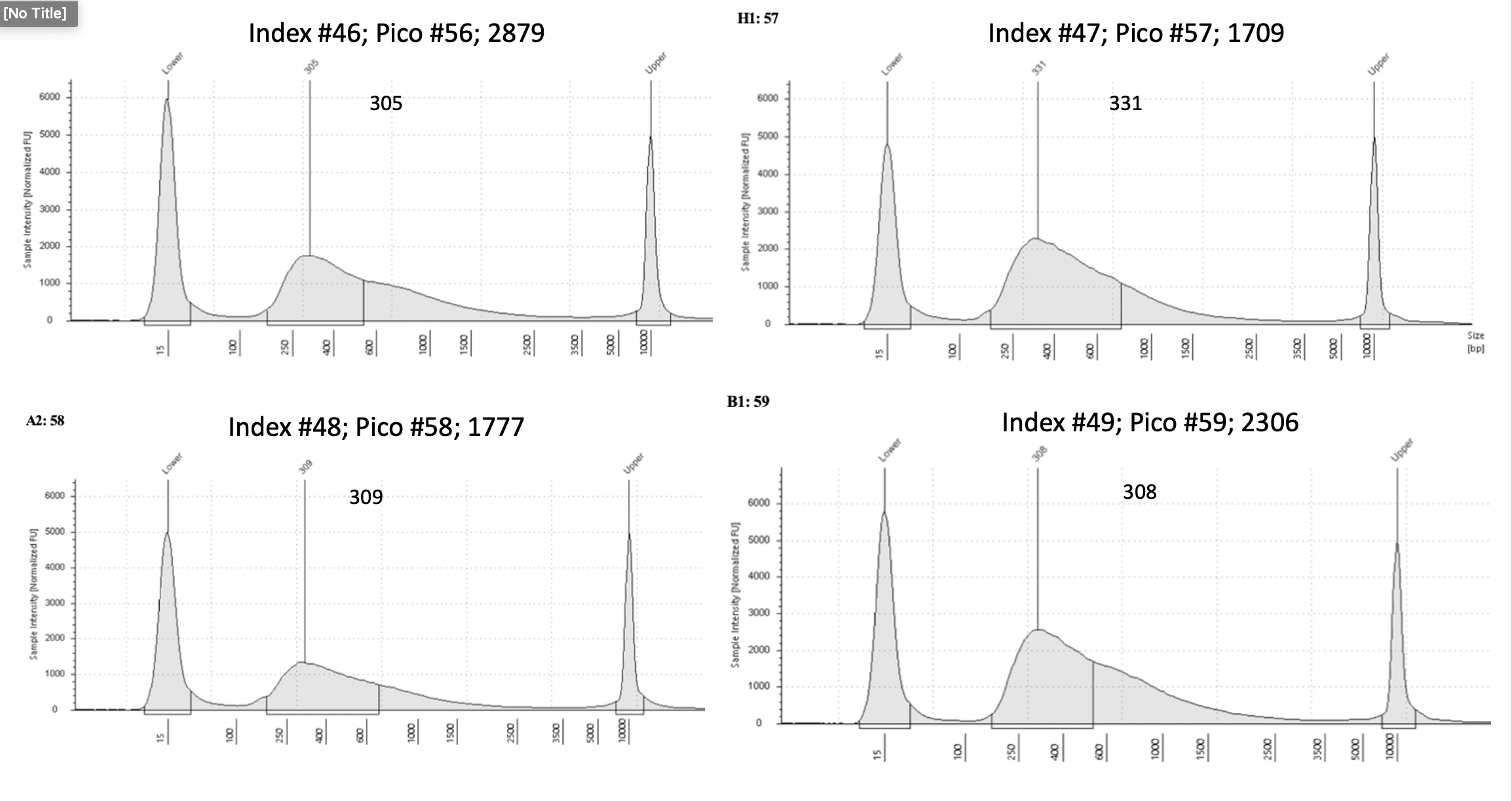
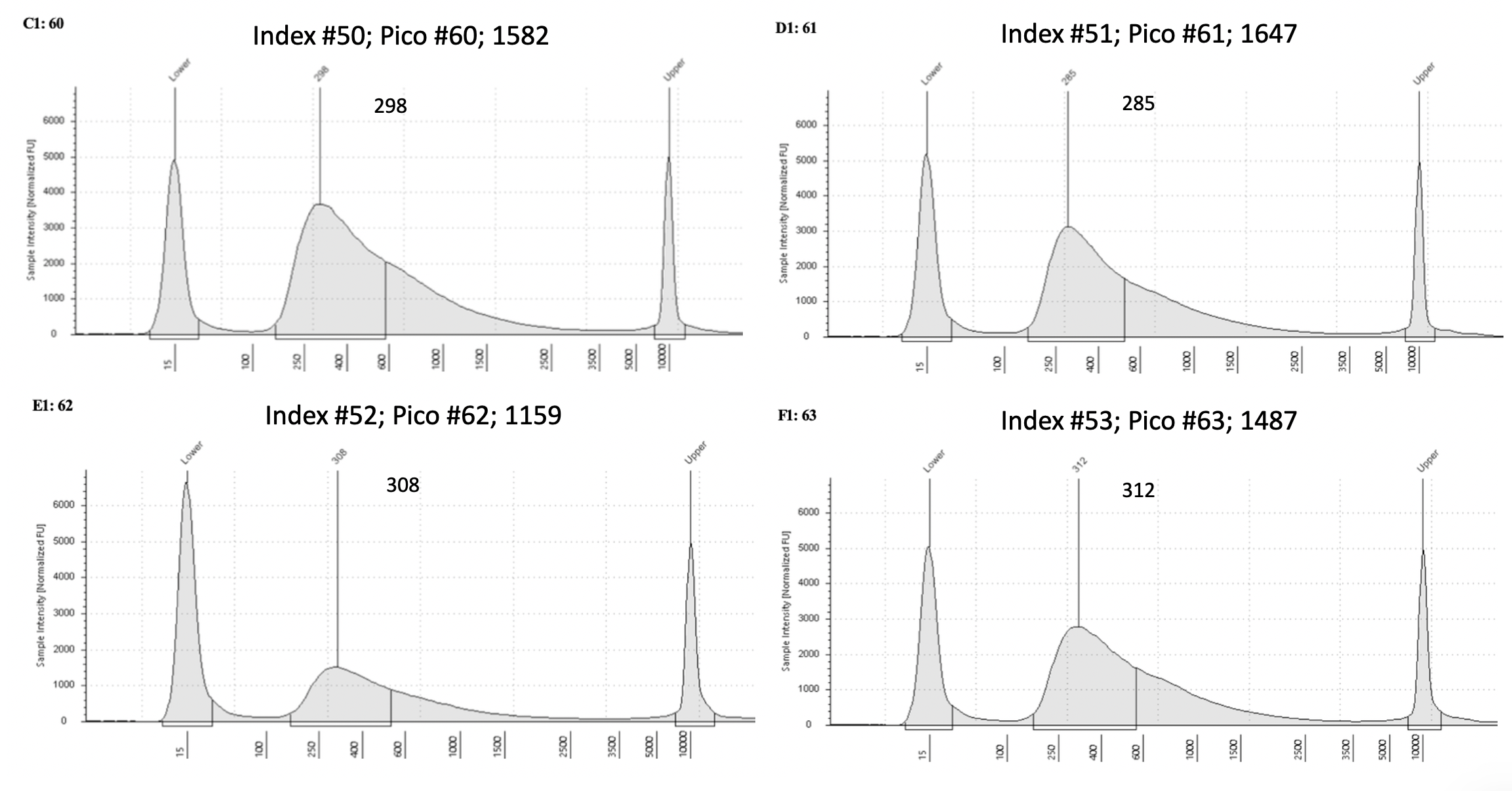
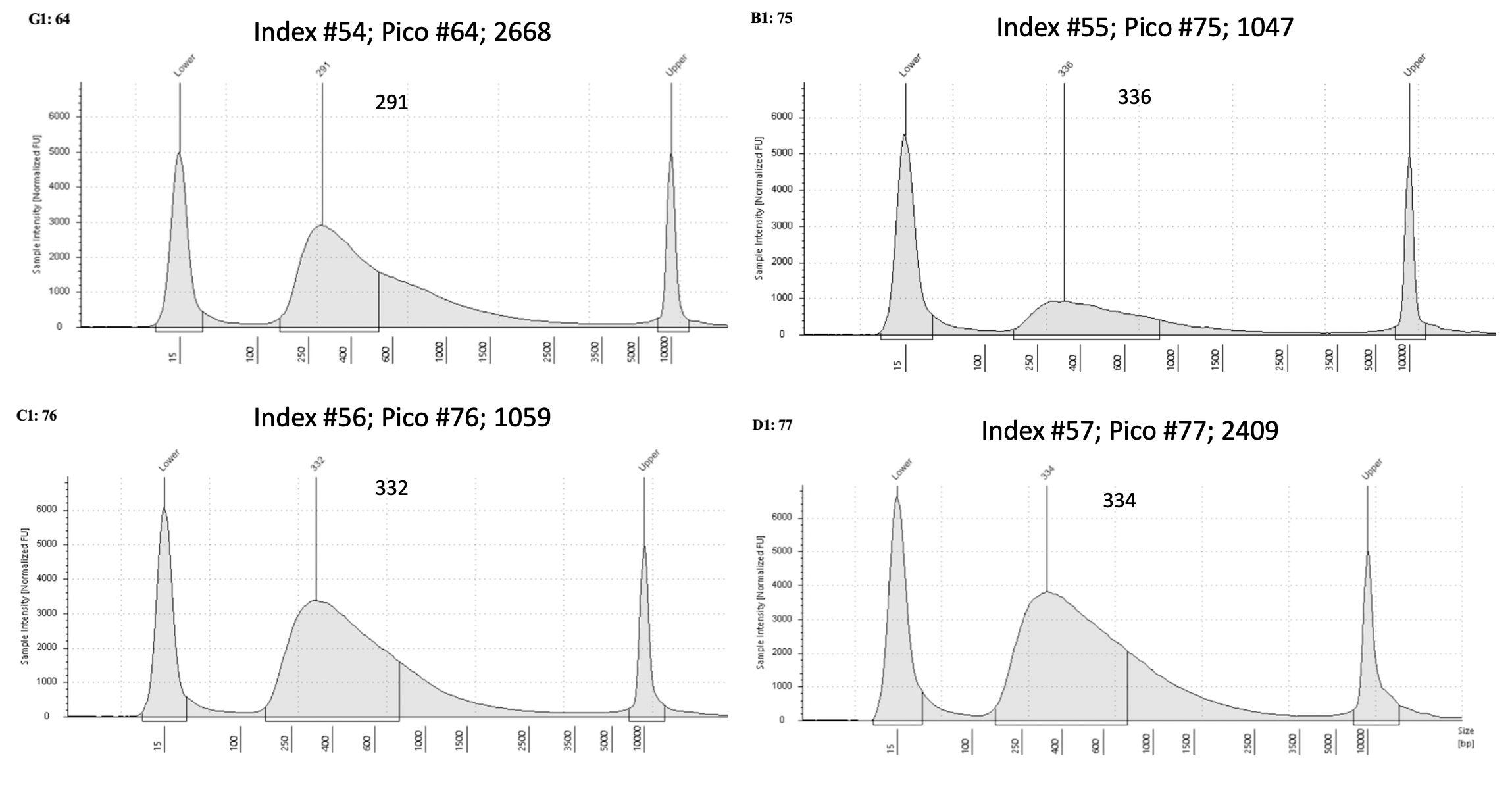
D5000 Tapestation results
Pico #1-3 tapestation here.
Pico #4-8 tapestation here.
Pico #9-18 tapestation here.
7 and 8 reamped tapestation here.
Pico #19-26 tapestation here.
Pico #27-42 TapeStation here.
Re-amps for Pico #27-42 tapestation here.
Pico #43-50 tapestation here
Pico #51-58 tapestation here
Pico #59-73 tapestation here
Pico #74,27,30,38,44,49 tapestation here
Pico #75-81, 72 tapestation here
Final D5000 Tapestation for all samples (59)
To view each image closer up, click on the image and this will open another tab with that image enlarged.