Becker DNA RNA Extractions
Danielle Becker Coral Samples Processing January 2020
DNA RNA Extractions
20200123 Test DNA RNA extraction round for Danielle Becker coral samples.
DNA/RNA Extractions from Danielle Becker’s Pocillopora coral samples using the following protocol: Zymo Duet RNA DNA Extraction Protocol and modified adult fragment tissue preparation steps described below:
- Half of coral fragment clipped and 500 μl of DNA/RNA shield from sample tube placed in tube with 2 mL 0.5 mm glass beads.
- 40 seconds at 20 Hz in the Tissue Lyser to remove tissue from coral skeleton.
- Supernatant removed and placed in 1.5 mL microcentrifuge tube for a stock solution (~700 μl).
- 500 μl of tissue homogenate, 50 μl of Proteinase K Digestion buffer (1:10 ratio with tissue homogenate volume), and 25 μl of Proteinase K added to a new 1.5 mL microcentrifuge tube.
- 575 μl of Lysis Buffer added (1:1 ratio to tissue, Proteinase K Digestion buffer, and Proteinase K volume)
- 1,150 μl of 100% molecular grade ethanol added to tissue for RNA portion of the protocol (1:1 ratio)
- 50 μl warmed Tris added to yellow spin column and incubated at room temperature for 5 minutes before centrifuged, and repeated for a total of 100 μl DNA.
- 50 μl warmed DNA RNA free water added to green spin column and incubated at room temperature for 5 minutes before centrifuged, and repeated for a total of 100 μl RNA.
Qubit used to check DNA and RNA quantity (ng/μl).
TapeStation used to check RNA quality.
Broad Range:
DNA Standard 1: 141.84 ng/μl
DNA Standard 2: 18,643.22 ng/μl
High Sensitivity:
RNA Standard 1: 50.93 ng/μl
RNA Standard 2: 963.31 ng/μl
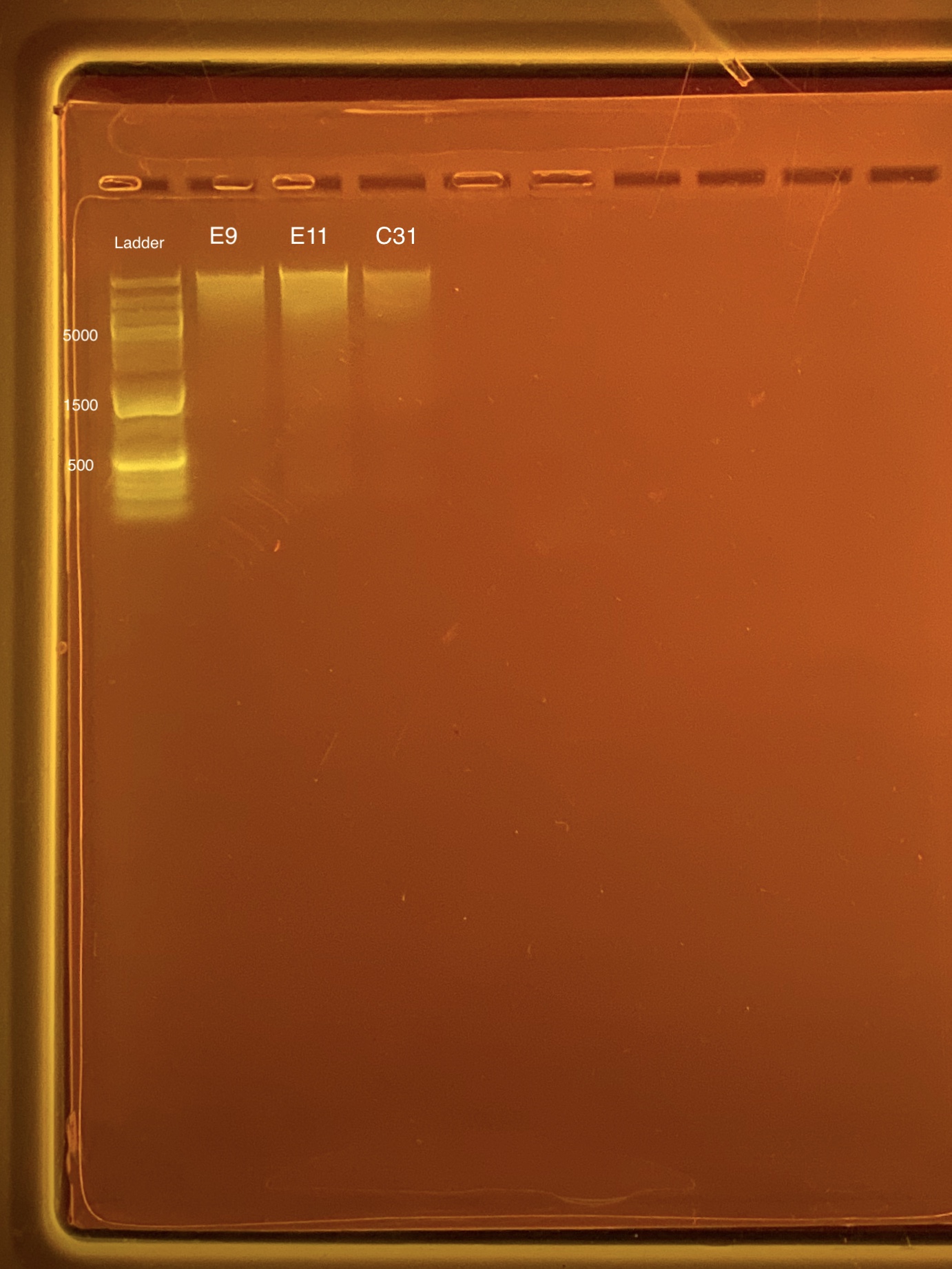
| Coral ID | DNA 1 | DNA 2 | Average DNA | RNA 1 | RNA 2 | Average RNA | RIN |
|---|---|---|---|---|---|---|---|
| E9 | 11.4 | 11.4 | 11.4 | 21.2 | 21.2 | 21.2 | 9.2 |
| E11 | 6.84 | 6.8 | 6.82 | 9.82 | 9.96 | 9.89 | 8.8 |
| C31 | 15.5 | 15.4 | 15.45 | 10.2 | 10.2 | 10.2 | 7.7 |
Link to 20200123 TapeStation report
20200127 DNA RNA extraction round for Danielle Becker coral samples.
DNA/RNA Extractions from Danielle Becker’s Pocillopora coral samples using the following protocol: Zymo Duet RNA DNA Extraction Protocol and modified adult fragment tissue preparation steps described below:
- Whole coral clipping and 700 μl of DNA/RNA shield from sample tube placed in tube with 2 mL 0.5 mm glass beads and 300 μl fresh DNA/RNA shield.
- 40 seconds at 20 Hz in the Tissue Lyser to remove tissue from coral skeleton.
- Supernatant removed and placed in 1.5 mL microcentrifuge tube for a stock solution (~700 μl).
- 500 μl of tissue homogenate, 50 μl of Proteinase K Digestion buffer (1:10 ratio with tissue homogenate volume), and 25 μl of Proteinase K added to a new 1.5 mL microcentrifuge tube.
- 575 μl of Lysis Buffer added (1:1 ratio to tissue, Proteinase K Digestion buffer, and Proteinase K volume)
- 1,150 μl of 100% molecular grade ethanol added to tissue for RNA portion of the protocol (1:1 ratio)
- 50 μl warmed Tris added to yellow spin column and incubated at room temperature for 5 minutes before centrifuged, and repeated for a total of 100 μl DNA.
- 50 μl warmed DNA RNA free water added to green spin column and incubated at room temperature for 5 minutes before centrifuged, and repeated for a total of 100 μl RNA.
Half of RNA flow-through from samples C30, E4, and E10 thrown out accidentally. 575 μl of sample (original sample, Proteinase K Digestion Buffer, and Proteinase K) and 575 μl of 100% molecular grade ethanol used. Spun down twice in total. All other samples followed 1,150 μl of sample and 1,150 μl of 100% ethanol protocol steps.
Qubit used to check DNA and RNA quantity (ng/μl).
TapeStation used to check RNA quality.
Broad Range:
DNA Standard 1: 142.77 ng/μl
DNA Standard 2: 20,503.32 ng/μl
High Sensitivity:
RNA Standard 1: 51.31 ng/μl
RNA Standard 2: 1,010.38 ng/μl
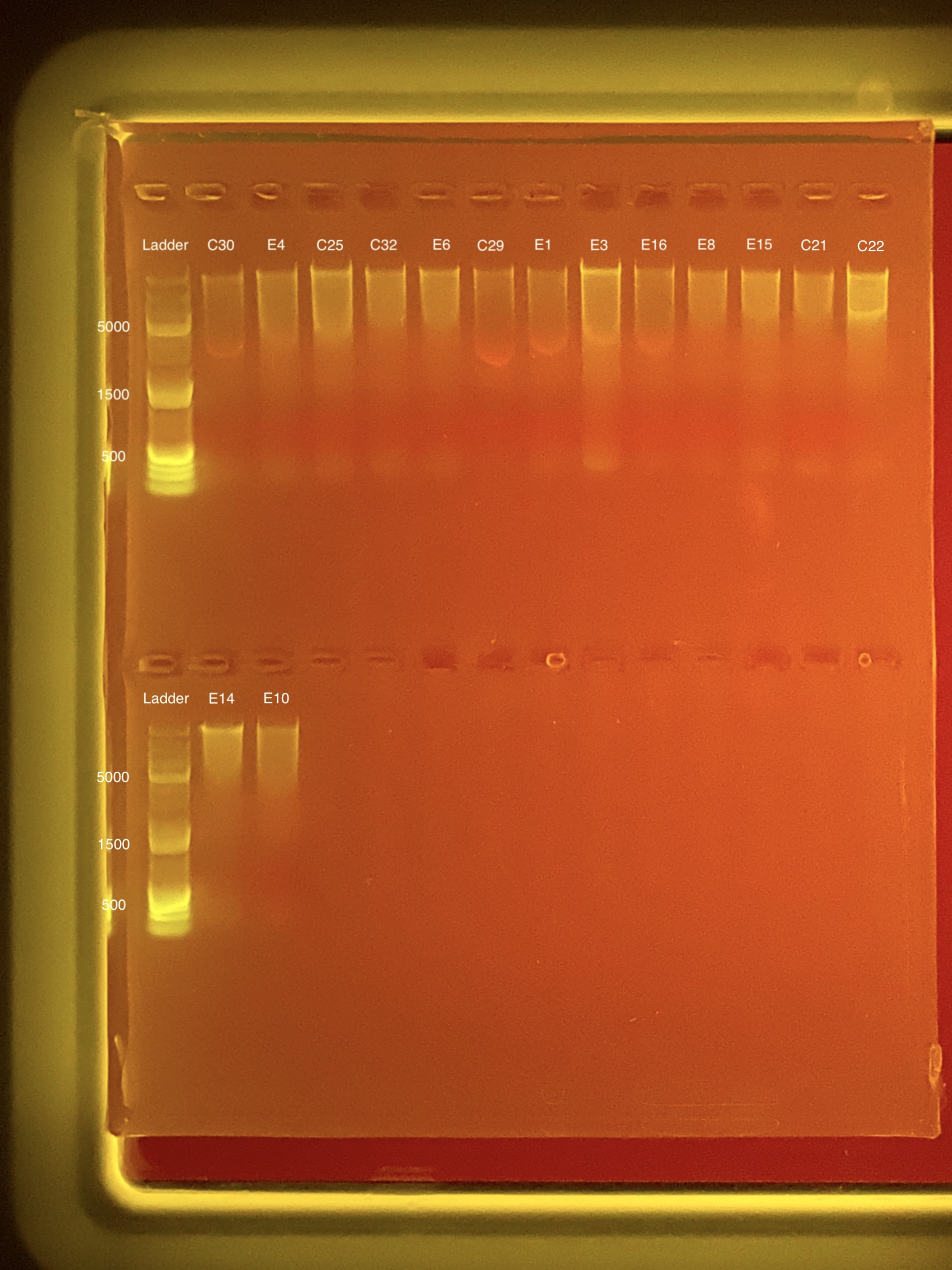
| Coral ID | DNA 1 | DNA 2 | Average DNA | RNA 1 | RNA 2 | Average RNA | RIN |
|---|---|---|---|---|---|---|---|
| C30 | 6.64 | 6.6 | 6.62 | ** | ** | ** | ** |
| E4 | 12 | 12 | 12 | 4.6 | 4.6 | 4.6 | ** |
| C25 | 19.7 | 19.7 | 19.7 | 18.3 | 18.3 | 18.3 | 9.2 |
| C32 | 7.72 | 7.68 | 7.7 | ** | ** | ** | ** |
| E6 | 8.34 | 8.32 | 8.33 | 8.38 | 8.44 | 8.41 | 9.1 |
| C29 | 7.38 | 7.36 | 7.37 | ** | ** | ** | ** |
| E1 | 6.08 | 6.04 | 6.06 | ** | ** | ** | ** |
| E3 | 23.8 | 23.8 | 23.8 | ** | ** | ** | ** |
| E16 | 12.4 | 12.4 | 12.4 | 7.76 | 7.88 | 7.82 | 9 |
| E8 | 4.68 | 4.66 | 4.67 | ** | ** | ** | ** |
| E15 | 6.56 | 6.52 | 6.54 | ** | ** | ** | ** |
| C21 | 6.2 | 6.18 | 6.19 | ** | ** | ** | ** |
| C22 | 14.1 | 14 | 14.05 | ** | ** | ** | ** |
| E14 | 9.58 | 9.52 | 9.55 | ** | ** | ** | ** |
| E10 | 9.86 | 9.82 | 9.84 | 4.4 | 4.4 | 4.4 | ** |
Link to 20200127 TapeStation report
20200128 DNA RNA extraction round for Danielle Becker coral samples.
DNA/RNA Extractions from Danielle Becker’s Pocillopora coral samples using the following protocol: Zymo Duet RNA DNA Extraction Protocol and modified adult fragment tissue preparation steps described below:
- Whole coral clipping and 700 μl of DNA/RNA shield from sample tube placed in tube with 2 mL 0.5 mm glass beads and 300 μl fresh DNA/RNA shield.
- 40 seconds at 20 Hz in the Tissue Lyser to remove tissue from coral skeleton.
- Supernatant removed and placed in 1.5 mL microcentrifuge tube for a stock solution (~700 μl).
- 600 μl of tissue homogenate, 60 μl of Proteinase K Digestion buffer (1:10 ratio with tissue homogenate volume), and 30 μl of Proteinase K added to a new 1.5 mL microcentrifuge tube.
- 690 μl of Lysis Buffer added (1:1 ratio to tissue, Proteinase K Digestion buffer, and Proteinase K volume)
- 1,380 μl of 100% molecular grade ethanol added to tissue for RNA portion of the protocol (1:1 ratio)
- 50 μl warmed Tris added to yellow spin column and incubated at room temperature for 5 minutes before centrifuged, and repeated for a total of 100 μl DNA.
- 50 μl warmed DNA RNA free water added to green spin column and incubated at room temperature for 5 minutes before centrifuged, and repeated for a total of 100 μl RNA.
Post tissue lyser: 1,000 μl DNA RNA Shield added to samples E5, C17, and E7 because there might be enough tissue on the skeleton to try another extraction if necessary. These samples had larger clippings.
Qubit used to check DNA and RNA quantity (ng/μl).
TapeStation used to check RNA quality.
Broad Range:
DNA Standard 1: 162.98 ng/μl
DNA Standard 2: 23,670.63 ng/μl
High Sensitivity:
RNA Standard 1: 51.25 ng/μl
RNA Standard 2: 995.63 ng/μl
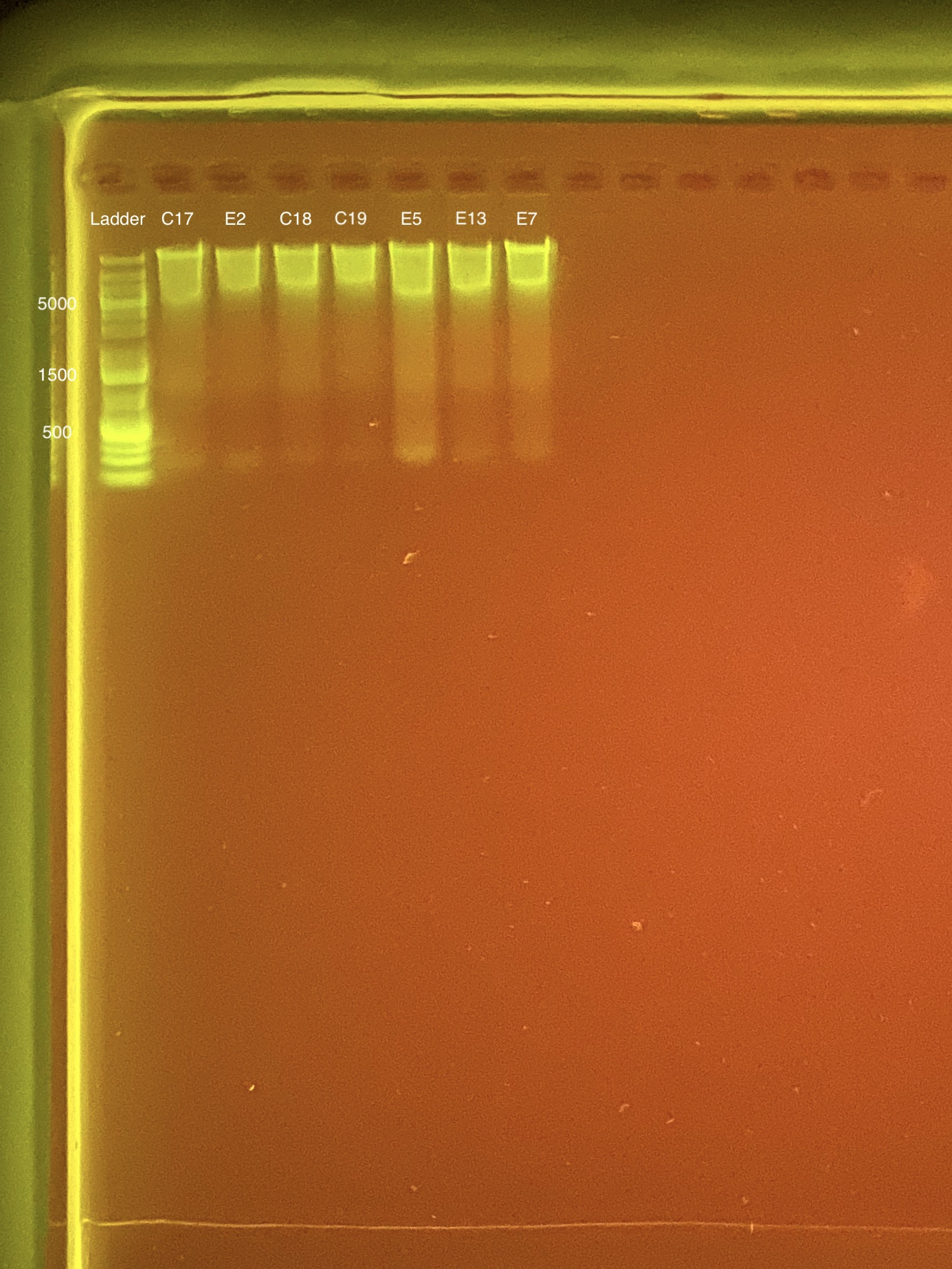
| Coral ID | DNA 1 | DNA 2 | Average DNA | RNA 1 | RNA 2 | Average RNA | RIN |
|---|---|---|---|---|---|---|---|
| C17 | 19.1 | 18.3 | 18.7 | 23 | 23 | 23 | 9.3 |
| E2 | 11.3 | 11.3 | 11.3 | 11 | 11.1 | 11.05 | 8.8 |
| C18 | 14.7 | 14.7 | 14.7 | 17.5 | 17.5 | 17.5 | 9.2 |
| C19 | 9.2 | 9.16 | 9.18 | 8.52 | 8.46 | 8.49 | 8.2 |
| E5 | 17.7 | 17.7 | 17.7 | 11.8 | 11.9 | 11.85 | 8.3 |
| E13 | 15.9 | 15.8 | 15.85 | 15.1 | 15 | 15.05 | 9.6 |
Link to 20200128 TapeStation report
Photos of clipping size to compare to qubit values:
Control 17:
Enriched 2:

Control 18:

Control 19:
Enriched 5:

Enriched 13:
Enriched 7:
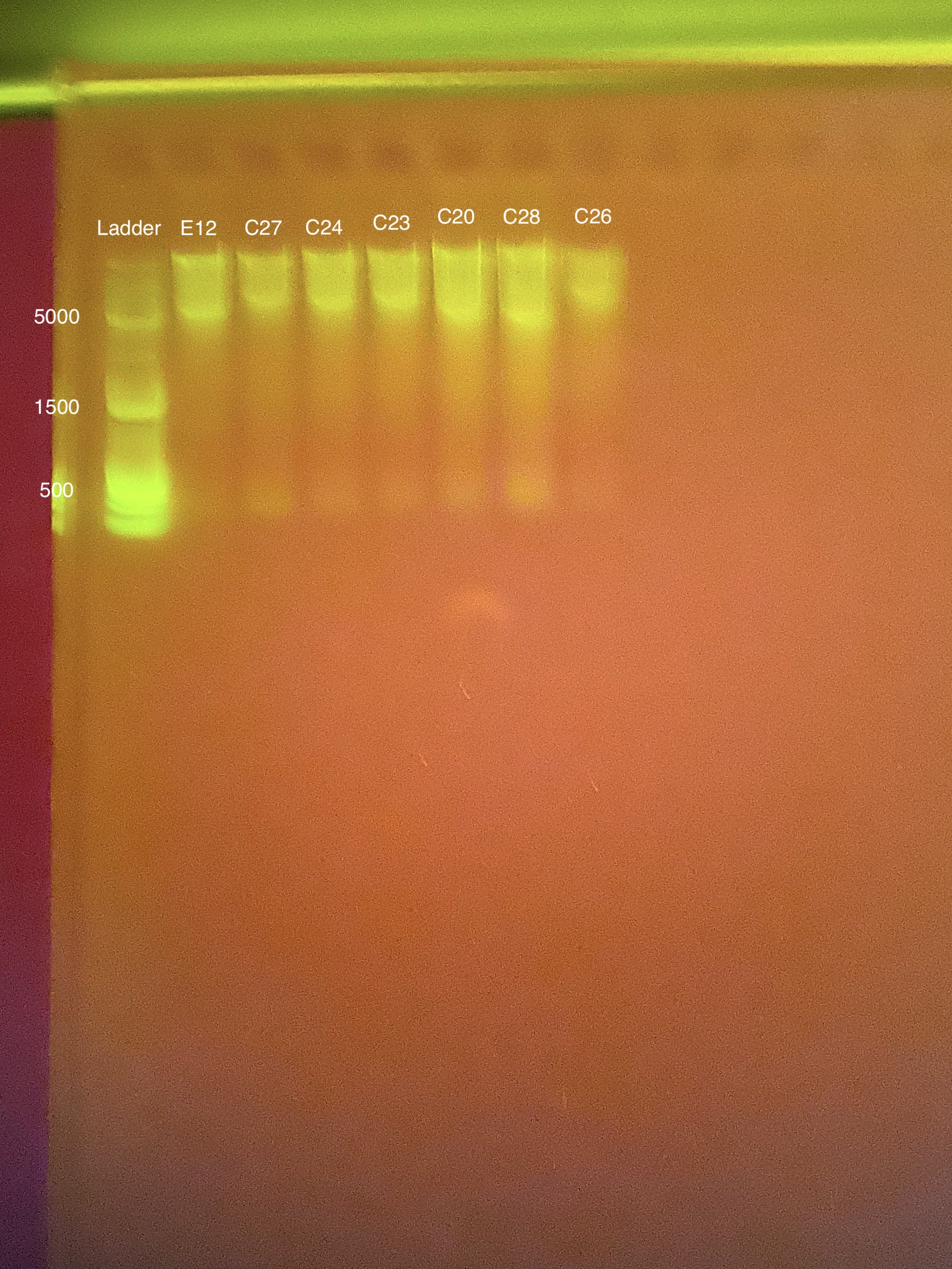
20200129 DNA RNA extraction round for Danielle Becker coral samples.
DNA/RNA Extractions from Danielle Becker’s Pocillopora coral samples using the following protocol: Zymo Duet RNA DNA Extraction Protocol and modified adult fragment tissue preparation steps described below:
- Whole coral clipping and 700 μl of DNA/RNA shield from sample tube placed in tube with 2 mL 0.5 mm glass beads and 300 μl fresh DNA/RNA shield.
- 40 seconds at 20 Hz in the Tissue Lyser to remove tissue from coral skeleton.
- Supernatant removed and placed in 1.5 mL microcentrifuge tube for a stock solution (~700 μl).
- 600 μl of tissue homogenate, 60 μl of Proteinase K Digestion buffer (1:10 ratio with tissue homogenate volume), and 30 μl of Proteinase K added to a new 1.5 mL microcentrifuge tube.
- 690 μl of Lysis Buffer added (1:1 ratio to tissue, Proteinase K Digestion buffer, and Proteinase K volume)
- 1,380 μl of 100% molecular grade ethanol added to tissue for RNA portion of the protocol (1:1 ratio)
- 50 μl warmed Tris added to yellow spin column and incubated at room temperature for 5 minutes before centrifuged, and repeated for a total of 100 μl DNA.
- 50 μl warmed DNA RNA free water added to green spin column and incubated at room temperature for 5 minutes before centrifuged, and repeated for a total of 100 μl RNA.
Post tissue lyser: 1,000 μl DNA RNA Shield added to larger sample clippings because there might be enough tissue on the skeleton to try another extraction if necessary. These samples had larger clippings.
1,830 μl of 100% ethanol accidentally added to all of the RNA samples instead of 1,380 μl.
Qubit used to check DNA and RNA quantity (ng/μl).
TapeStation used to check RNA quality.
Broad Range:
DNA Standard 1: 155.65 ng/μl
DNA Standard 2: 22,158.47 ng/μl
High Sensitivity:
RNA Standard 1: 52.72 ng/μl
RNA Standard 2: 1,037.95 ng/μl
| Date | Coral ID | DNA 1 | DNA 2 | Average DNA (ng_uL) | RNA 1 | RNA 2 | Average RNA (ng_uL) | RIN |
|---|---|---|---|---|---|---|---|---|
| 20200129 | E12 | 17.6 | 17.5 | 17.55 | 13.9 | 13.9 | 13.9 | 9.1 |
| 20200129 | C20 | 21 | 20.8 | 20.9 | 11.7 | 11.7 | 11.7 | 8.2 |
| 20200129 | C23 | 10.9 | 10.9 | 10.9 | 6.4 | 6.36 | 6.38 | 7.4 |
| 20200129 | C24 | 12.7 | 12.7 | 12.7 | 13.5 | 13.6 | 13.55 | 9.3 |
| 20200129 | C26 | 7.94 | 7.92 | 7.93 | ** | ** | ** | 6 |
| 20200129 | C27 | 9.18 | 9.16 | 9.17 | ** | ** | ** | ** |
| 20200129 | C28 | 22.2 | 22.2 | 22.2 | 12.2 | 12.2 | 12.2 | 8.9 |
Link to 20200129 TapeStation report
Summary of DNA RNA Extraction Results
| Date | Coral ID | DNA 1 | DNA 2 | Average DNA (ng_uL) | RNA 1 | RNA 2 | Average RNA (ng_uL) | RIN |
|---|---|---|---|---|---|---|---|---|
| 20200127 | E1 | 6.08 | 6.04 | 6.06 | ** | ** | ** | ** |
| 20200128 | E2 | 11.3 | 11.3 | 11.3 | 11 | 11.1 | 11.05 | 8.8 |
| 20200127 | E3 | 23.8 | 23.8 | 23.8 | ** | ** | ** | ** |
| 20200127 | E4 | 12 | 12 | 12 | 4.6 | 4.6 | 4.6 | ** |
| 20200128 | E5 | 17.7 | 17.7 | 17.7 | 11.8 | 11.9 | 11.85 | 8.3 |
| 20200127 | E6 | 8.34 | 8.32 | 8.33 | 8.38 | 8.44 | 8.41 | 9.1 |
| 20200128 | E7 | 18.9 | 18.9 | 18.9 | 15.6 | 15.5 | 15.55 | 8.8 |
| 20200127 | E8 | 4.68 | 4.66 | 4.67 | ** | ** | ** | ** |
| 20200123 | E9 | 11.4 | 11.4 | 11.4 | 21.2 | 21.2 | 21.2 | 9.2 |
| 20200127 | E10 | 9.86 | 9.82 | 9.84 | 4.4 | 4.4 | 4.4 | ** |
| 20200123 | E11 | 6.84 | 6.8 | 6.82 | 9.82 | 9.96 | 9.89 | 8.8 |
| 20200129 | E12 | 17.6 | 17.5 | 17.55 | 13.9 | 13.9 | 13.9 | 9.1 |
| 20200128 | E13 | 15.9 | 15.8 | 15.85 | 15.1 | 15 | 15.05 | 9.6 |
| 20200127 | E14 | 9.58 | 9.52 | 9.55 | ** | ** | ** | ** |
| 20200127 | E15 | 6.56 | 6.52 | 6.54 | ** | ** | ** | ** |
| 20200127 | E16 | 12.4 | 12.4 | 12.4 | 7.76 | 7.88 | 7.82 | 9 |
| 20200128 | C17 | 19.1 | 18.3 | 18.7 | 23 | 23 | 23 | 9.3 |
| 20200128 | C18 | 14.7 | 14.7 | 14.7 | 17.5 | 17.5 | 17.5 | 9.2 |
| 20200128 | C19 | 9.2 | 9.16 | 9.18 | 8.52 | 8.46 | 8.49 | 8.2 |
| 20200129 | C20 | 21 | 20.8 | 20.9 | 11.7 | 11.7 | 11.7 | 8.2 |
| 20200127 | C21 | 6.2 | 6.18 | 6.19 | ** | ** | ** | ** |
| 20200127 | C22 | 14.1 | 14 | 14.05 | ** | ** | ** | ** |
| 20200129 | C23 | 10.9 | 10.9 | 10.9 | 6.4 | 6.36 | 6.38 | 7.4 |
| 20200129 | C24 | 12.7 | 12.7 | 12.7 | 13.5 | 13.6 | 13.55 | 9.3 |
| 20200127 | C25 | 19.7 | 19.7 | 19.7 | 18.3 | 18.3 | 18.3 | 9.2 |
| 20200129 | C26 | 7.94 | 7.92 | 7.93 | ** | ** | ** | 6 |
| 20200129 | C27 | 9.18 | 9.16 | 9.17 | ** | ** | ** | ** |
| 20200129 | C28 | 22.2 | 22.2 | 22.2 | 12.2 | 12.2 | 12.2 | 8.9 |
| 20200127 | C29 | 7.38 | 7.36 | 7.37 | ** | ** | ** | ** |
| 20200127 | C30 | 6.64 | 6.6 | 6.62 | ** | ** | ** | ** |
| 20200123 | C31 | 15.5 | 15.4 | 15.45 | 10.2 | 10.2 | 10.2 | 7.7 |
| 20200127 | C32 | 7.72 | 7.68 | 7.7 | ** | ** | ** | ** |
ITS2 Sequencing
The Internal Transcribed Spacer 2 (ITS2) gene was isolated from all DNA samples above using the following Putnam Lab ITS2 Sequencing Protocol.
Dilution aliquot calculations:
| Amplicon ID | Coral ID | Extraction Date | Hard DNA (ng_ul) | DNA for dilution (ul) | Water for dilution (ul) | Dilution Strip Tube # |
|---|---|---|---|---|---|---|
| 1 | E9 | 20200127 | 11.4 | 2.89 | 7.11 | 1 |
| 2 | E11 | 20200128 | 6.82 | 4.84 | 5.16 | 1 |
| 3 | C31 | 20200127 | 15.45 | 2.14 | 7.86 | 1 |
| 4 | C30 | 20200127 | 6.62 | 4.98 | 5.02 | 1 |
| 5 | E4 | 20200128 | 12 | 2.75 | 7.25 | 1 |
| 6 | C25 | 20200127 | 19.7 | 1.68 | 8.32 | 1 |
| 7 | C32 | 20200128 | 7.7 | 4.29 | 5.71 | 1 |
| 8 | E6 | 20200127 | 8.33 | 3.96 | 6.04 | 1 |
| 9 | C29 | 20200123 | 7.37 | 4.48 | 5.52 | 2 |
| 10 | E1 | 20200127 | 6.06 | 5.45 | 4.55 | 2 |
| 11 | E3 | 20200123 | 23.8 | 1.39 | 8.61 | 2 |
| 12 | E16 | 20200129 | 12.4 | 2.66 | 7.34 | 2 |
| 13 | E8 | 20200128 | 4.67 | 7.07 | 2.93 | 2 |
| 14 | E15 | 20200127 | 6.54 | 5.05 | 4.95 | 2 |
| 15 | C21 | 20200127 | 6.19 | 5.33 | 4.67 | 2 |
| 16 | C22 | 20200127 | 14.05 | 2.35 | 7.65 | 2 |
| 17 | E14 | 20200128 | 9.55 | 3.46 | 6.54 | 3 |
| 18 | E10 | 20200128 | 9.84 | 3.35 | 6.65 | 3 |
| 19 | C17 | 20200128 | 18.7 | 1.76 | 8.24 | 3 |
| 20 | E2 | 20200129 | 11.3 | 2.92 | 7.08 | 3 |
| 21 | C18 | 20200127 | 14.7 | 2.24 | 7.76 | 3 |
| 22 | C19 | 20200127 | 9.18 | 3.59 | 6.41 | 3 |
| 23 | E5 | 20200129 | 17.7 | 1.86 | 8.14 | 3 |
| 24 | E13 | 20200129 | 15.85 | 2.08 | 7.92 | 3 |
| 25 | E7 | 20200127 | 18.9 | 1.75 | 8.25 | 4 |
| 26 | E12 | 20200129 | 17.55 | 1.88 | 8.12 | 4 |
| 27 | C27 | 20200129 | 9.17 | 3.6 | 6.4 | 4 |
| 28 | C24 | 20200129 | 12.7 | 2.6 | 7.4 | 4 |
| 29 | C23 | 20200127 | 10.9 | 3.03 | 6.97 | 4 |
| 30 | C20 | 20200127 | 20.9 | 1.58 | 8.42 | 4 |
| 31 | C28 | 20200123 | 22.2 | 1.49 | 8.51 | 4 |
| 32 | C26 | 20200127 | 7.93 | 4.16 | 5.84 | 4 |
| 33 | MIX | 18.2 | 4.57 | 20.43 | 5 | |
| 34 | NEG CON | 12.75 | 6.53 | 18.47 | 5 |
96-well plate maps:
Plate 1:
| NA | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 1 | 1 | 1 | 9 | 9 | 9 | 17 | 17 | 17 | NEGCON | NEGCON | NEGCON |
| B | 2 | 2 | 2 | 10 | 10 | 10 | 18 | 18 | 18 | |||
| C | 3 | 3 | 3 | 11 | 11 | 11 | 19 | 19 | 19 | |||
| D | 4 | 4 | 4 | 12 | 12 | 12 | 20 | 20 | 20 | |||
| E | 5 | 5 | 5 | 13 | 13 | 13 | 21 | 21 | 21 | |||
| F | 6 | 6 | 6 | 14 | 14 | 14 | 22 | 22 | 22 | |||
| G | 7 | 7 | 7 | 15 | 15 | 15 | 23 | 23 | 23 | |||
| H | 8 | 8 | 8 | 16 | 16 | 16 | 24 | 24 | 24 |
Plate 2:
| NA | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 25 | 25 | 25 | 33 | 33 | 33 | ||||||
| B | 26 | 26 | 26 | NEGCON | NEGCON | NEGCON | ||||||
| C | 27 | 27 | 27 | |||||||||
| D | 28 | 28 | 28 | |||||||||
| E | 29 | 29 | 29 | |||||||||
| F | 30 | 30 | 30 | |||||||||
| G | 31 | 31 | 31 | |||||||||
| H | 32 | 32 | 32 |
Gel Electrophoresis Image:
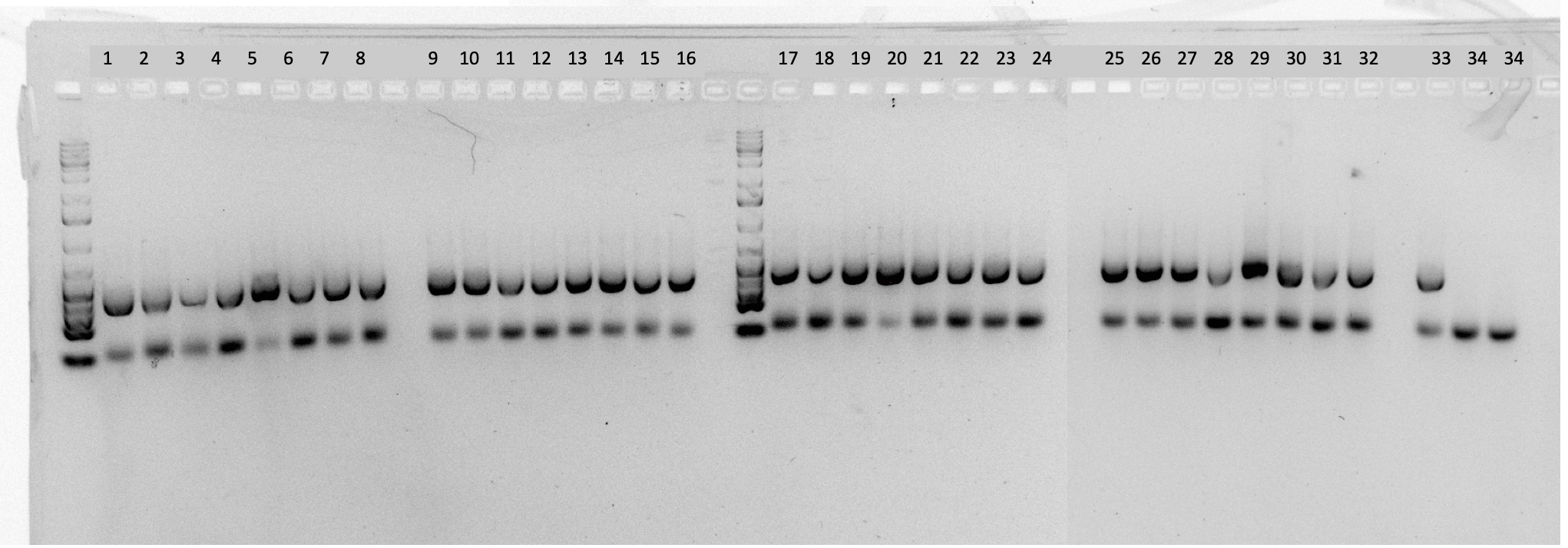
Sequencing center ID:
| DataID | AmpliconID | SampleName | AmpliconGene | StripTube Number |
|---|---|---|---|---|
| HP401 | 1 | E9 | ITS2 | 1 |
| HP402 | 2 | E11 | ITS2 | 1 |
| HP403 | 3 | C31 | ITS2 | 1 |
| HP404 | 4 | C30 | ITS2 | 1 |
| HP405 | 5 | E4 | ITS2 | 1 |
| HP406 | 6 | C25 | ITS2 | 1 |
| HP407 | 7 | C32 | ITS2 | 1 |
| HP408 | 8 | E6 | ITS2 | 1 |
| HP409 | 9 | C29 | ITS2 | 2 |
| HP410 | 10 | E1 | ITS2 | 2 |
| HP411 | 11 | E3 | ITS2 | 2 |
| HP412 | 12 | E16 | ITS2 | 2 |
| HP413 | 13 | E8 | ITS2 | 2 |
| HP414 | 14 | E15 | ITS2 | 2 |
| HP415 | 15 | C21 | ITS2 | 2 |
| HP416 | 16 | C22 | ITS2 | 2 |
| HP417 | 17 | E14 | ITS2 | 3 |
| HP418 | 18 | E10 | ITS2 | 3 |
| HP419 | 19 | C17 | ITS2 | 3 |
| HP420 | 20 | E2 | ITS2 | 3 |
| HP421 | 21 | C18 | ITS2 | 3 |
| HP422 | 22 | C19 | ITS2 | 3 |
| HP423 | 23 | E5 | ITS2 | 3 |
| HP424 | 24 | E13 | ITS2 | 3 |
| HP425 | 25 | E7 | ITS2 | 4 |
| HP426 | 26 | E12 | ITS2 | 4 |
| HP427 | 27 | C27 | ITS2 | 4 |
| HP428 | 28 | C24 | ITS2 | 4 |
| HP429 | 29 | C23 | ITS2 | 4 |
| HP430 | 30 | C20 | ITS2 | 4 |
| HP431 | 31 | C28 | ITS2 | 4 |
| HP432 | 32 | C26 | ITS2 | 4 |
| HP433 | 33 | MIX | ITS2 | 5 |
| HP434 | 34 | NEG CON | ITS2 | 5 |
Re-extracting the samples with no RNA yield
Extractions from re-bead beating coral fragments + 300 uL of original shield
DNA/RNA Extractions from Danielle Becker’s Pocillopora coral samples using the following protocol: Zymo Duet RNA DNA Extraction Protocol and modified adult fragment tissue preparation steps described below:
Tissue fragment preparation:
- Thawing the bead tube that contains the fragments and beads from the original extraction
- Adding 300 uL of the original DNA RNA shield and 700 uL of new DNA RNA shield
- 40 seconds at 20 Hz in the Tissue Lyser to remove reamining tissue from coral skeleton
- Supernatant removed and placed in 1.5 mL microcentrifuge tube for the extraction protocol (Zymo link above) with the following modifications.
- No stock solution leftover from 20200901 extraction. ~300 uL of stock solution remains from the original extraction in case 20200901 extraction fails.
Extraction modification steps:
- 700 μl of tissue homogenate, 70 μl of Proteinase K Digestion buffer (1:10 ratio with tissue homogenate volume), and 35 μl of Proteinase K added to a new 1.5 mL microcentrifuge tube.
- 805 μl of Lysis Buffer added (1:1 ratio to tissue, Proteinase K Digestion buffer, and Proteinase K volume)
- 1,610 μl of 100% molecular grade ethanol added to tissue for RNA portion of the protocol (1:1 ratio)
20200901 notes:
- No incubation step used for final DNA elution steps (accidentally skipped). Lower DNA yield from these samples but should still be usable.
20200907 notes:
- Supernatant is very clear, hardly any tissue on these fragment pieces. Worried about not getting enough genomic pool from the coral. Starting to think the original stock would better biologically even though there is only 300 uL leftover.
- C32 did not have any original DNA RNA shield left. 1,000 uL of new RNA DNA shield used.
Qubit and TapeStation Results
Extractions from coral fragment pieces and original DNA RNA shield
| Date | Coral ID | DNA 1 | DNA 2 | Average DNA (ng_uL) | RNA 1 | RNA 2 | Average RNA (ng_uL) | RIN | Pass? |
|---|---|---|---|---|---|---|---|---|---|
| 20200901 | E3 | 10.9 | 10.8 | 10.85 | 16.8 | 16.8 | 16.8 | 8.3 | Yes |
| 20200901 | E8 | 3 | 2.98 | 2.99 | 6.96 | 7 | 6.98 | ** | No |
| 20200901 | E14 | 7.46 | 7.4 | 7.43 | 15.5 | 15.5 | 15.5 | 8.9 | Yes |
| 20200901 | C22 | 7.78 | 7.76 | 7.77 | 23.6 | 23.6 | 23.6 | 9.4 | Yes |
| 20200907 | E1 | 9.5 | 9.46 | 9.48 | 8.74 | 8.72 | 8.73 | ** | Re do tape? |
| 20200907 | E15 | 3.74 | 3.68 | 3.71 | 5.54 | 5.42 | 5.48 | ** | No |
| 20200907 | C21 | 2.66 | 2.64 | 2.65 | 7.72 | 7.82 | 7.77 | 8.8 | Yes |
| 20200907 | C26 | 4.78 | 4.74 | 4.76 | 5.88 | 5.9 | 5.89 | ** | No |
| 20200907 | C27 | 3.52 | 3.5 | 3.51 | 4.2 | 4.4 | 4.3 | ** | No |
| 20200907 | C29 | 4.5 | 4.48 | 4.49 | 6.74 | 6.72 | 6.73 | ** | No |
| 20200907 | C30 | 5.34 | 5.28 | 5.31 | 9.3 | 9.32 | 9.31 | 8.6 | Yes |
| 20200907 | C32 | 3.02 | 3.02 | 3.02 | ** | ** | ** | ** | No |
| 20200915 | E4 | 5.2 | 4.96 | 5.08 | 15.1 | 15.1 | 15.1 | 9.3 | Yes |
| 20200915 | E10 | 11.4 | 11.3 | 11.35 | 23.2 | 23.2 | 8 | 8 | Yes |
20200901 Extractions:
DNA BR Standard 1: 199.24 ng/μl
DNA BR Standard 2: 22,582.06 ng/μl
RNA HS Standard 1: 49.18 ng/μl
RNA HS Standard 2: 907.40 ng/μl
20200907 Extractions:
DNA BR Standard 1: 192.94 ng/μl
DNA BR Standard 2: 21,521.22 ng/μl
RNA HS Standard 1: 50.71 ng/μl
RNA HS Standard 2: 952.91 ng/μl
2020915 Extractions:
DNA BR Standard 1: 168.02 ng/μl
DNA BR Standard 2: 19,640.60 ng/μl
RNA HS Standard 1: 50.14 ng/μl
RNA HS Standard 2: 924.90 ng/μl
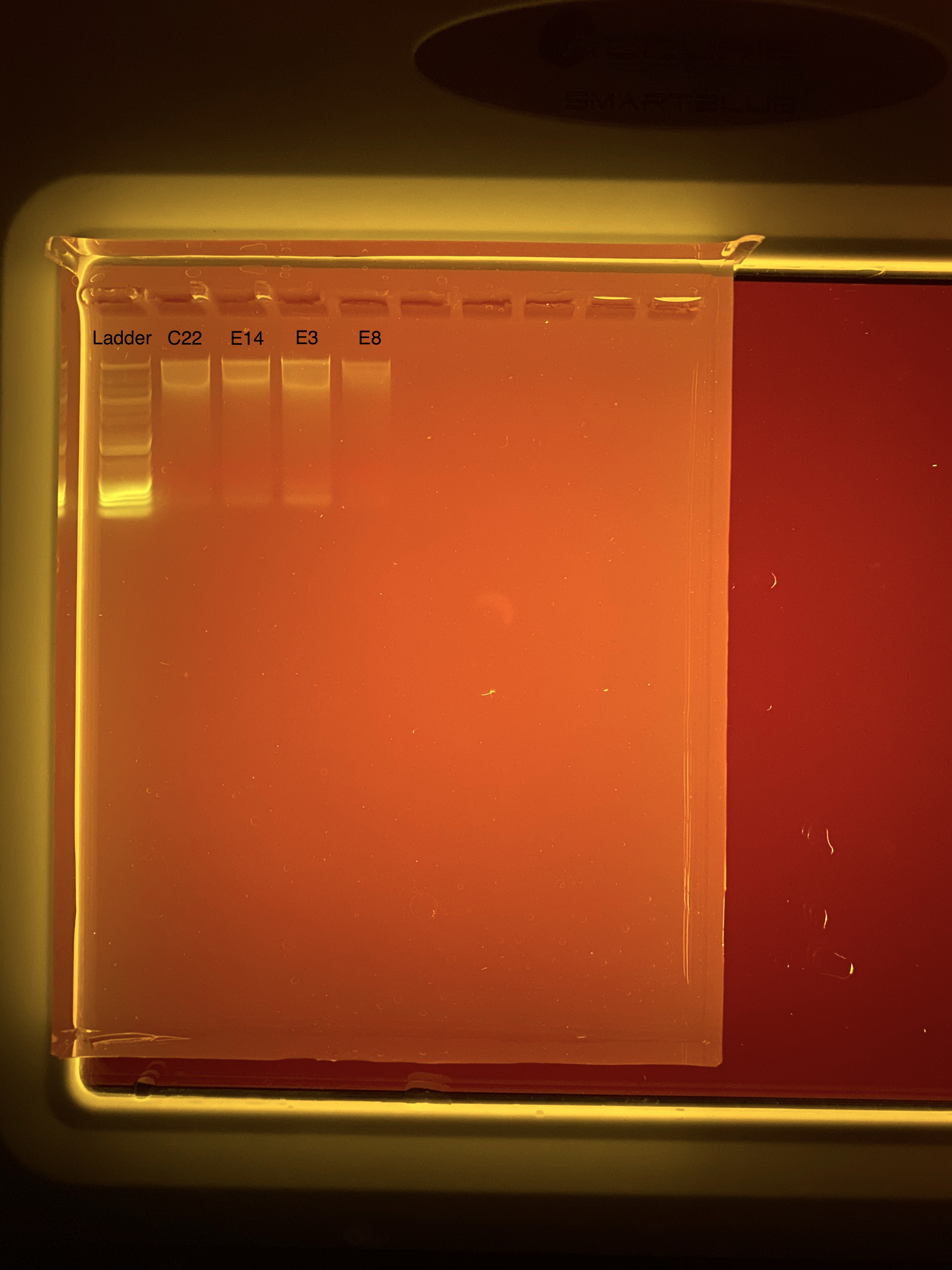
20200901 E3, E8, E14, C22 TapeStation Results
20200901 E3, E8, E14, C22 Gel Image
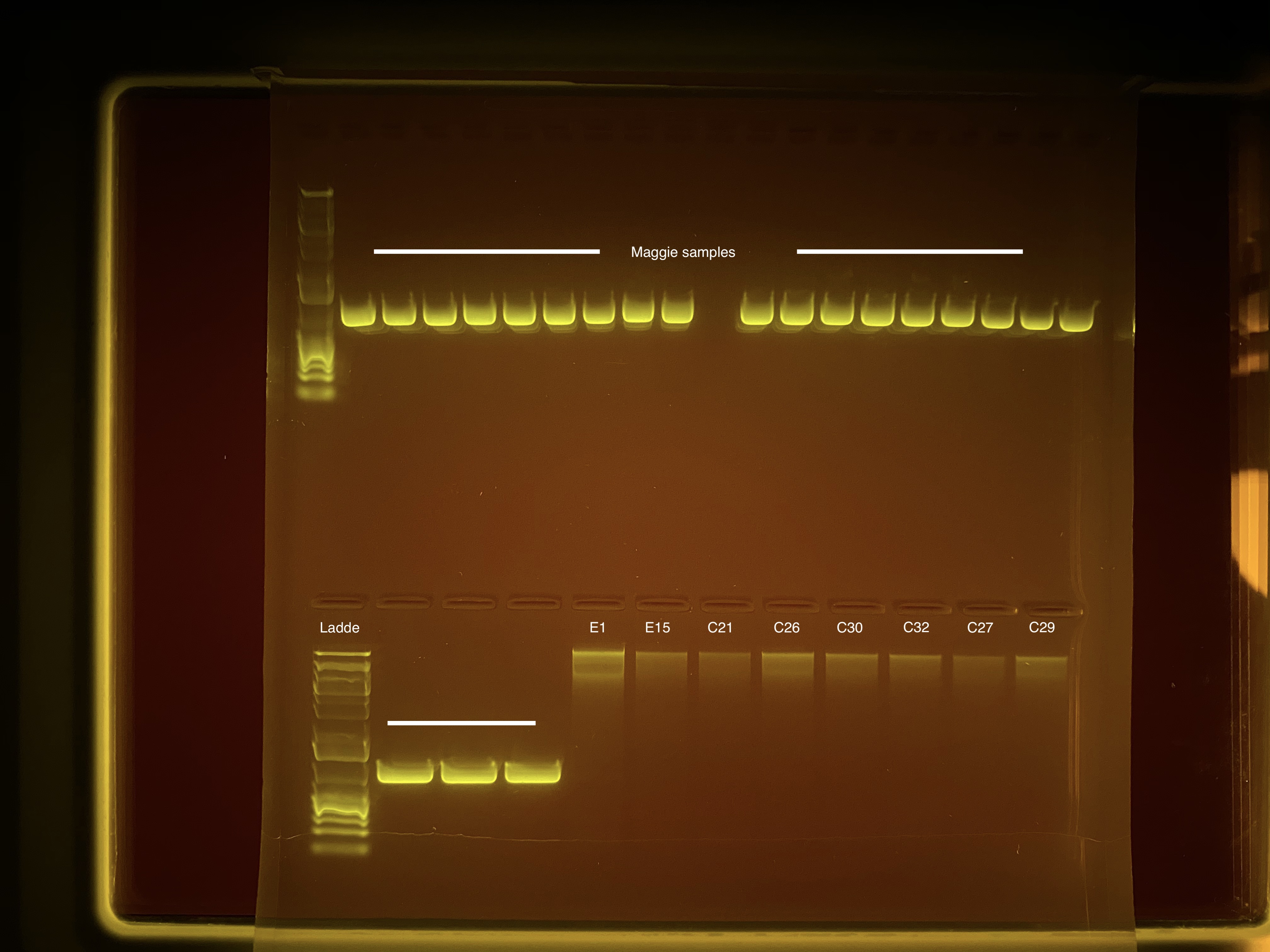
20200908 TapeStation Results
20200908 Gel Image
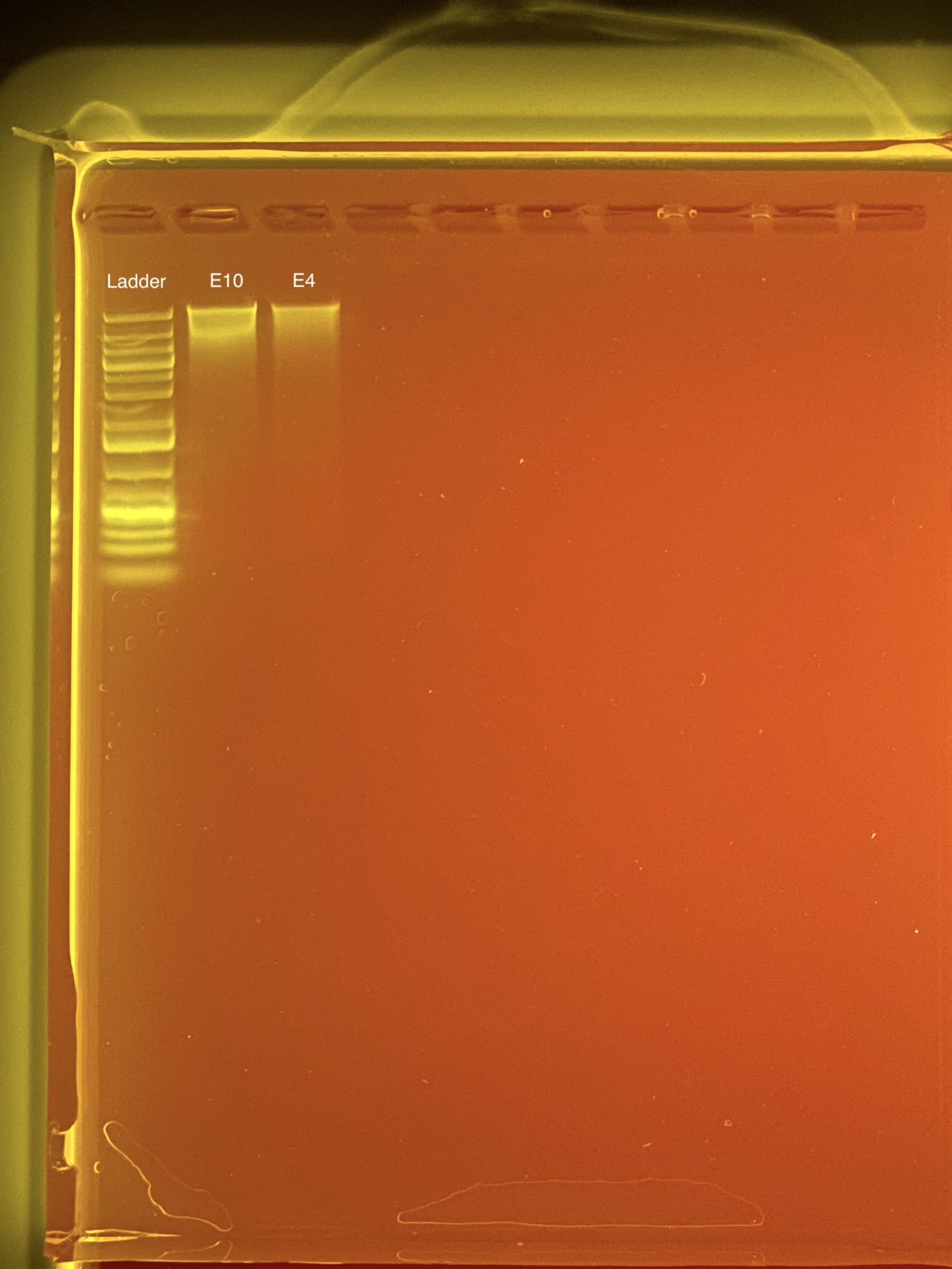
20200915 TapeStation
20200915 Gel image
Although I did get RNA from this re-extraction round, I am now thinking that doing extractions on the original stock (only 300 uL) would be better biologically. The original stock would have more coral tissue and is darker in color than what I got from beating the coral fragments again. Only issue is that I wasn’t originally able to get enough from 300 uL on the first time around which is why I hadn’t done re-extractions on those yet. If this works, I would take the RNA and DNA from the stock instead of what was done on 20200901 and 20200907.
Next steps: 20200912 Extractions on the 300 uL stock from all samples. Two rounds of 6 samples. This is the last shot on this batch of corals. If still not satisfied with results, then consider doing cites for the batch in Mo’orea.
Extractions from 300 uL original stock solution from the first time bead beating the fragments
20200911:
2 rounds of extractions: 1.) E8, C22, E14, E8, C29, E15 and 2.) C30, C21, E1, C27, C26, C32
Stock is from original Tissue Lyser run, go straight into ProK Buffer and ProK step. (300 uL sample, 30 uL Prok Buffer, 15 uL ProK).
20200914 Qubit, TapeStation
DNA BR Standard 1: 194.38 ng/μl
DNA BR Standard 2: 21,798.00 ng/μl
RNA HS Standard 1: 49.37 ng/μl
RNA HS Standard 2: 941.70 ng/μl
| Date | Coral ID | DNA 1 | DNA 2 | Average DNA (ng_uL) | RNA 1 | RNA 2 | Average RNA (ng_uL) | RIN | Pass? |
|---|---|---|---|---|---|---|---|---|---|
| 20200911 | E3 | 8.6 | 8.56 | 8.58 | 7.28 | 7.36 | 7.32 | ** | No |
| 20200911 | E8 | ** | ** | ** | ** | ** | ** | ** | No |
| 20200911 | E14 | 5.04 | 5.02 | 5.03 | 5.12 | 5.1 | 5.11 | ** | No |
| 20200911 | C22 | 7.9 | 7.86 | 7.88 | 10.7 | 10.7 | 10.7 | 8.9 | Yes |
| 20200911 | E1 | 2.66 | 2.64 | 2.65 | 5.18 | 5.26 | 5.22 | ** | No |
| 20200911 | E15 | ** | ** | ** | ** | ** | ** | ** | No |
| 20200911 | C21 | 2.44 | 2.42 | 2.43 | 4.6 | 4.6 | 4.6 | ** | No |
| 20200911 | C26 | ** | ** | ** | ** | ** | ** | ** | No |
| 20200911 | C27 | ** | ** | ** | ** | ** | ** | ** | No |
| 20200911 | C29 | 2.56 | 2.56 | 2.56 | 5.14 | 5.08 | 5.11 | ** | No |
| 20200911 | C30 | 2.42 | 2.4 | 2.41 | 6.04 | 6.04 | 6.04 | ** | No |
| 20200911 | C32 | 2.5 | 2.46 | 2.48 | ** | ** | ** | ** | No |
20200914 TapeStation Results
No gel image for these extractions because GelGreen was accidentally left out of the gel. Not re-done because Qubit values were low anyways.
This method didn’t work as well as I had hoped. Could be that the stock de-thawing caused degradation of DNA RNA? Use the extractions done with re-bead beating the coral fragments.
Just to double check:
Re-qubit C26 RNA from January 2019.
RNA HS Standard 1: 49.57 ng/μl
RNA HS Standard 2: 964.11 ng/μl
Value 1: 5.04 ng/uL Value 2: 5.04 ng/uL
Re-TapeStation E1 from January 2019.
Value returned as too low to detect. TapeStation Report
Removed E1 and C26 from the final list to send to sequencing.
20200921:
The below will be combined and re-qubit prior to sequencing.
| Date | Coral ID | DNA 1 | DNA 2 | Average DNA (ng_uL) | RNA 1 | RNA 2 | Average RNA (ng_uL) | RIN | Pass? | Qualitative RIN by HP | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 20200907 | E1 | 9.5 | 9.46 | 9.48 | 8.74 | 8.72 | 8.73 | ** | No | Fine | Combine |
| 20200911 | E1 | 2.66 | 2.64 | 2.65 | 5.18 | 5.26 | 5.22 | ** | No | Fine | Combine |
| 20200129 | C26 | 7.94 | 7.92 | 7.93 | 5.04 | 5.04 | 5.04 | 6 | No | OK | Combine |
| 20200907 | C26 | 4.78 | 4.74 | 4.76 | 5.88 | 5.9 | 5.89 | ** | No | OK | Combine |
| 20200907 | C29 | 4.5 | 4.48 | 4.49 | 6.74 | 6.72 | 6.73 | ** | No | Fine | Combine |
| 20200911 | C29 | 2.56 | 2.56 | 2.56 | 5.14 | 5.08 | 5.11 | ** | No | Fine | Combine |
| 20200907 | C32 | 3.02 | 3.02 | 3.02 | ** | ** | ** | ** | No | Fine, but low | Combine |
| 20200911 | C32 | 2.5 | 2.46 | 2.48 | ** | ** | ** | ** | No | Fine, but low | Combine |
New qubit values:
| Date | Coral ID | DNA 1 | DNA 2 | Average DNA (ng_uL) | RNA 1 | RNA 2 | Average RNA (ng_uL) | RIN | Pass? | Qualitative RIN by HP |
|---|---|---|---|---|---|---|---|---|---|---|
| 20200922 | E1 | 7.6 | 7.5 | 7.55 | 9.52 | 9.6 | 9.56 | 8.6 | Yes | Fine |
| 20200922 | C26 | 7.52 | 7.44 | 7.48 | 6.3 | 6.34 | 6.32 | ** | Yes | Fine |
| 20200922 | C29 | 4.84 | 4.78 | 4.81 | 6.44 | 6.42 | 6.43 | ** | Yes | Fine |
| 20200922 | C32 | 3.6 | 3.48 | 3.54 | 4.4 | ** | 4.4 | ** | Yes | Fine |
Combined DNA on gel 20200923:
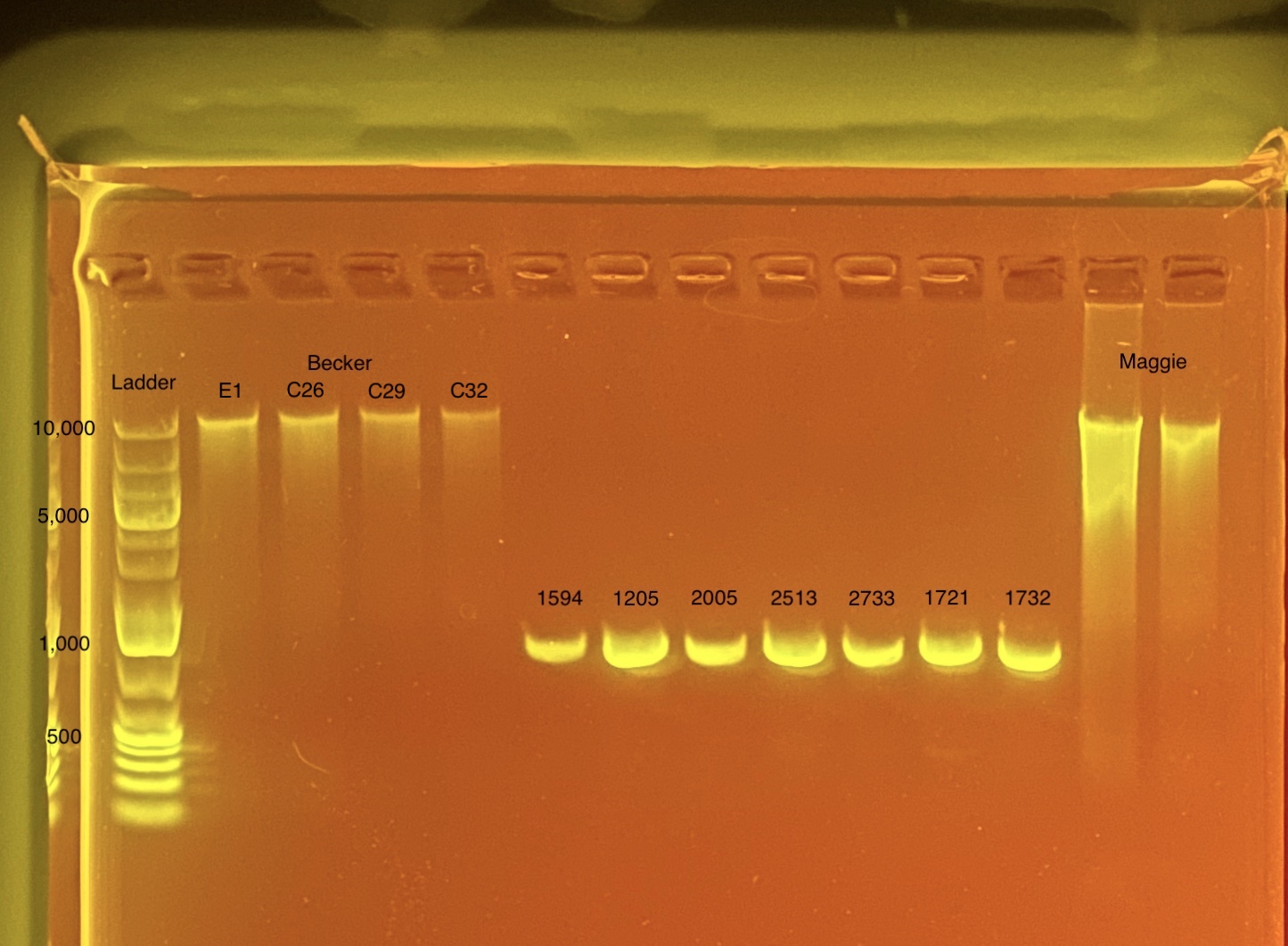
Final Extraction Values
20200916 final thoughts: Some of the coral had enough tissue on the fragments to be re-bead beated successfully. There was also likely DNA and RNA in the original shield volume. I now think this is the better option biologically because freeze-thawing stock or tissue homogenate more than once degrades the DNA and RNA in the homogenate. Which you can see based on the results of the second type of re-extractions based on the stock, and this also happened to Kevin (labmate) with his 2017 vs 2018 tissue homogenates.
Dates in January 2020 are original extraction values. Dates in September 2020 are re-extraction values.
Below are the samples that have passed DNA and RNA quality and quantity tested post extraction. All fragments are together in a box in the new, blue -80C freezer.
| Date | Coral ID | DNA 1 | DNA 2 | Average DNA (ng_uL) | RNA 1 | RNA 2 | Average RNA (ng_uL) | RIN | Pass? | Qualitative RIN by HP |
|---|---|---|---|---|---|---|---|---|---|---|
| 20200128 | C17 | 19.1 | 18.3 | 18.7 | 23 | 23 | 23 | 9.3 | Yes | |
| 20200128 | C18 | 14.7 | 14.7 | 14.7 | 17.5 | 17.5 | 17.5 | 9.2 | Yes | |
| 20200128 | C19 | 9.2 | 9.16 | 9.18 | 8.52 | 8.46 | 8.49 | 8.2 | Yes | |
| 20200129 | C20 | 21 | 20.8 | 20.9 | 11.7 | 11.7 | 11.7 | 8.2 | Yes | |
| 20200907 | C21 | 2.66 | 2.64 | 2.65 | 7.72 | 7.82 | 7.77 | 8.8 | Yes | |
| 20200901 | C22 | 7.78 | 7.76 | 7.77 | 23.6 | 23.6 | 23.6 | 9.4 | Yes | |
| 20200129 | C23 | 10.9 | 10.9 | 10.9 | 6.4 | 6.36 | 6.38 | 7.4 | Yes | |
| 20200129 | C24 | 12.7 | 12.7 | 12.7 | 13.5 | 13.6 | 13.55 | 9.3 | Yes | |
| 20200127 | C25 | 19.7 | 19.7 | 19.7 | 18.3 | 18.3 | 18.3 | 9.2 | Yes | |
| 20200922 | C26 | 7.52 | 7.44 | 7.48 | 6.3 | 6.34 | 6.32 | ** | Yes | Fine |
| 20200907 | C27 | 3.52 | 3.5 | 3.51 | 4.2 | 4.4 | 4.3 | ** | Yes | Fine |
| 20200129 | C28 | 22.2 | 22.2 | 22.2 | 12.2 | 12.2 | 12.2 | 8.9 | Yes | |
| 20200922 | C29 | 4.84 | 4.78 | 4.81 | 6.44 | 6.42 | 6.43 | ** | Yes | Fine |
| 20200907 | C30 | 5.34 | 5.28 | 5.31 | 9.3 | 9.32 | 9.31 | 8.6 | Yes | |
| 20200123 | C31 | 15.5 | 15.4 | 15.45 | 10.2 | 10.2 | 10.2 | 7.7 | Yes | |
| 20200922 | C32 | 3.6 | 3.48 | 3.54 | 4.4 | *** | 4.4 | ** | Yes | Fine |
| 20200922 | E1 | 7.6 | 7.5 | 7.55 | 9.52 | 9.6 | 9.56 | 8.6 | Yes | Fine |
| 20200128 | E2 | 11.3 | 11.3 | 11.3 | 11 | 11.1 | 11.05 | 8.8 | Yes | |
| 20200901 | E3 | 10.9 | 10.8 | 10.85 | 16.8 | 16.8 | 16.8 | 8.3 | Yes | |
| 20200915 | E4 | 5.2 | 4.96 | 5.08 | 15.1 | 15.1 | 15.1 | 9.3 | Yes | |
| 20200128 | E5 | 17.7 | 17.7 | 17.7 | 11.8 | 11.9 | 11.85 | 8.3 | Yes | |
| 20200127 | E6 | 8.34 | 8.32 | 8.33 | 8.38 | 8.44 | 8.41 | 9.1 | Yes | |
| 20200128 | E7 | 18.9 | 18.9 | 18.9 | 15.6 | 15.5 | 15.55 | 8.8 | Yes | |
| 20200901 | E8 | 3 | 2.98 | 2.99 | 6.96 | 7 | 6.98 | ** | Yes | Fine |
| 20200123 | E9 | 11.4 | 11.4 | 11.4 | 21.2 | 21.2 | 21.2 | 9.2 | Yes | |
| 20200915 | E10 | 11.4 | 11.3 | 11.35 | 23.2 | 23.2 | 8 | 8 | Yes | |
| 20200123 | E11 | 6.84 | 6.8 | 6.82 | 9.82 | 9.96 | 9.89 | 8.8 | Yes | |
| 20200129 | E12 | 17.6 | 17.5 | 17.55 | 13.9 | 13.9 | 13.9 | 9.1 | Yes | |
| 20200128 | E13 | 15.9 | 15.8 | 15.85 | 15.1 | 15 | 15.05 | 9.6 | Yes | |
| 20200901 | E14 | 7.46 | 7.4 | 7.43 | 15.5 | 15.5 | 15.5 | 8.9 | Yes | |
| 20200907 | E15 | 3.74 | 3.68 | 3.71 | 5.54 | 5.42 | 5.48 | ** | Yes | Fine |
| 20200127 | E16 | 12.4 | 12.4 | 12.4 | 7.76 | 7.88 | 7.82 | 9 | Yes |

- All four boxes contain saved material from the extractions. Once DNA and RNA is back from sequencing and backed up in several places, we can throw away most of the contents of the three white boxes. There are a couple of tubes with coral fragments that can be saved but we won’t need the rest.

- Tubes are also labeled on the sides with dates of extraction. Match these with the extraction dates in the tables above to find desired DNA tube.

Google spreadsheet with raw extraction data
mtORF amplication M. Schedl notebook post
- mtORF and ITS2 amplication was done with DNA from January 2020 extractions. This was completed prior to September 2020 re-extractions for DNA and RNA in the same genomic pool.
RNASeq Prep for Genewiz
Active spreadsheet with the below data.
I chose to send 500 ng because some samples had low quantity values and all samples sent to Genewiz need to have the same quantity of DNA. uL_to_500ng calculated by =500/Qubit. Check mark indicates the full volume needed was added. Two samples did not have enough total sample so that will be less 500 ng; we will send anyway.
| Date | Coral ID | RNA 1 | RNA 2 | Avg_RNA (ng/uL) | uL_to_500ng | Notes |
|---|---|---|---|---|---|---|
| 20200128 | C17 | 23 | 23 | 23 | 21.74 | ✓ |
| 20200128 | C18 | 17.5 | 17.5 | 17.5 | 28.57 | ✓ |
| 20200128 | C19 | 8.52 | 8.46 | 8.49 | 58.89 | ✓ |
| 20200129 | C20 | 11.7 | 11.7 | 11.7 | 42.74 | ✓ |
| 20200907 | C21 | 7.72 | 7.82 | 7.77 | 64.35 | ✓ |
| 20200901 | C22 | 23.6 | 23.6 | 23.6 | 21.19 | ✓ |
| 20200129 | C23 | 6.4 | 6.36 | 6.38 | 78.37 | ✓ |
| 20200129 | C24 | 13.5 | 13.6 | 13.55 | 36.90 | ✓ |
| 20200127 | C25 | 18.3 | 18.3 | 18.3 | 27.32 | ✓ |
| 20200129 | C28 | 12.2 | 12.2 | 12.2 | 40.98 | ✓ |
| 20200907 | C30 | 9.3 | 9.32 | 9.31 | 53.71 | ✓ |
| 20200123 | C31 | 10.2 | 10.2 | 10.2 | 49.02 | ✓ |
| 20200123 | E11 | 9.82 | 9.96 | 9.89 | 50.56 | ✓ |
| 20200129 | E12 | 13.9 | 13.9 | 13.9 | 35.97 | ✓ |
| 20200128 | E13 | 15.1 | 15 | 15.05 | 33.22 | ✓ |
| 20200901 | E14 | 15.5 | 15.5 | 15.5 | 32.26 | ✓ |
| 20200127 | E16 | 7.76 | 7.88 | 7.82 | 63.94 | ✓ |
| 20200128 | E2 | 11 | 11.1 | 11.05 | 45.25 | ✓ |
| 20200901 | E3 | 16.8 | 16.8 | 16.8 | 29.76 | ✓ |
| 20200915 | E4 | 15.1 | 15.1 | 15.1 | 33.11 | ✓ |
| 20200128 | E5 | 11.8 | 11.9 | 11.85 | 42.19 | ✓ |
| 20200127 | E6 | 8.38 | 8.44 | 8.41 | 59.45 | ✓ |
| 20200128 | E7 | 15.6 | 15.5 | 15.55 | 32.15 | ✓ |
| 20200123 | E9 | 21.2 | 21.2 | 21.2 | 23.58 | ✓ |
| 20200915 | E10 | 23.2 | 23.2 | 23.2 | 21.55 | ✓ |
| 20200901 | E8 | 6.96 | 7 | 6.98 | 71.63 | ✓ |
| 20200922 | E1 | 9.52 | 9.6 | 9.56 | 52.30 | ✓ |
| 20200922 | C26 | 6.3 | 6.34 | 6.32 | 79.11 | ✓ |
| 20200922 | C29 | 6.44 | 6.42 | 6.43 | 77.76 | ✓ |
| 20200922 | C32 | 4.4 | ** | 4.4 | 113.64 | ✓; had enough b/c combined 2 elutions |
| 20200907 | E15 | 5.54 | 5.42 | 5.48 | 91.24 | 84.8 uL added; that was all we had |
| 20200907 | C27 | 4.2 | 4.4 | 4.3 | 116.28 | 85.6 uL added; that was all we had |
20200923 Update: LN2 ordered; waiting for that to arrive prior to shipping. Samples are parafilmed and ready to be sent.
