16S V3V4 Sample Processing
Sample Processing for 16S V3V4 primers
16S PCR Protocol here.
Excel processing sheet here. I’m using excel and github instead of google sheets so that my processing sheets are version controlled.
Contents:
- Primers
- PCR settings
- Testing with 338F
- Testing with 341F
- Higher Annealing Temperature Test
- Sequencing test run 1
Primers
| Primer | GSC Adapter Overhang | Primer Sequence | Sequence to be ordered: Adapter + Primer Sequence | Total Length | Primer Length |
|---|---|---|---|---|---|
| 338F forward | TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG | ACTCCTACGGGAGGCAGCA | TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGACTCCTACGGGAGGCAGCA | 52 | 19 |
| 806RB reverse | GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG | GGACTACNVGGGTWTCTAAT | GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGGACTACNVGGGTWTCTAAT | 54 | 20 |
| 341F forward | TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG | CCTACGGGNGGCWGCAG | TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG | 50 | 17 |
| 515F forward | TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG | GTGCCAGCMGCCGCGGTAA | TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGTGCCAGCMGCCGCGGTAA | 52 | 19 |
- 338F order sheet from IDT
- 806RB order sheet from IDT
- 341F order sheet from IDT
- 515F order sheet from IDT
We received a concentrate of primer and needed to resuspend the 338F primer. Resuspending concenrate to 100 uM = started with 82.6 nM so IDT sheet said to add 826 uL to create 100 uM. Our stock will be 100 uM and then I diluted a portion to 10 uM for use in PCR protocol.
V3/V4 amplicon size: 380–400 bp + 50-60 bp for primer & adapter overhang (410-460 bp)
Dilutions to make 10 uM primer
C1 x V1 = C2 x V2 100 uM * X = 10 uM * 200 uL X = 2,000 / 100 X = 20 uL
200 uL of 10 uM primer = 20 uL 200 uM + 180 uL Ultra Pure water
PCR settings
PCR program used for V4 primers: 515F and 806RB
| Temperature | Time | Repeat |
|---|---|---|
| 95 °C | 2 min | 1 |
| *95 °C | 20 s | 35 |
| *57 °C | 15s | |
| *72 °C | 5 min | |
| 72 °C | 10 min | 1 |
PCR program used for V3/V4 primers: 338F/341F and 806RB
| Temperature | Time | Repeat |
|---|---|---|
| 95 °C | 2 min | 1 |
| *95 °C | 20 s | x30 |
| *57 °C | 30 s | |
| *72 °C | 20 s | |
| 72 °C | 5 min | 1 |
Testing with 338F
20220321 Trial 1
2 Pacuta, 2 Mcap adults, 2 eggs, 2 settled recruits and 1 negative control and 1 positive control (Kevin’s). We’re using Kevin’s as a positive control because in our V4 region sequencing, his samples had low host amplification compared to mine and Ariana’s. In this test, if all the bands are the same size (Kevin, mine and Ariana’s) then either all of them amplified the same targets (either all microbiome or all host - goal is the microbiome).
| Strip tube # | Plug ID | Species | Life stage | Project | Dilution plate | Well | Concentration | Notes |
|---|---|---|---|---|---|---|---|---|
| 1 | 2878 | P. acuta | Adult | HoloInt | DILUTIONS PLATE 2 - 5 TP RUN | A11 | 4 ng/uL | |
| 2 | 2513 | P. acuta | Adult | HoloInt | DILUTIONS PLATE 2 - 5 TP RUN | B3 | 4 ng/uL | 2513 for 55C/56C; 1184 (B4) for 57C |
| 3 | ES-17 (M-212) | M. capitata | Adult | KBay | SAMPLE # PLATE 1 | A12 | 4 ng/uL | ES-17 for 55C/56C; ES-18 for 57C |
| 4 | ES-16 (M-217) | M. capitata | Adult | KBay | SAMPLE # PLATE 1 | H11 | 4 ng/uL | |
| 5 | AH-9 D1 (TP1) | M. capitata | Eggs | A. Huffmyer | SAMPLE # PLATE 1 | E7 | 4 ng/uL | |
| 6 | AH-10 D2 (TP1) | M. capitata | Eggs | A. Huffmyer | SAMPLE # PLATE 1 | F7 | 4 ng/uL | |
| 7 | AH-2 (Plug_10 48 hps) | M. capitata | Settled recruits | A. Huffmyer | SAMPLE # PLATE 1 | F6 | 4 ng/uL | |
| 8 | AH-3 (Plug_4 48 hps) | M. capitata | Settled recruits | A. Huffmyer | SAMPLE # PLATE 1 | G7 | 4 ng/uL | |
| 9 | KW-10 (R37-A1) | P. asteroides | Adult | KW_PJB | SAMPLE # PLATE 1 | A1 | 4 ng/uL | |
| 10 | NA | NA | NA | Negative control | NA | NA | NA |
| Master Mix Calculations | |||
|---|---|---|---|
| uL | N (3031.05) | uL | |
| Phusion MM | 12.5 | 94.5 | 1181.25 |
| F primer | 0.5 | 94.5 | 47.25 |
| R primer | 0.5 | 94.5 | 47.25 |
| Water | 10.5 | 94.5 | 992.25 |
| Trial # | Temperature | n= |
|---|---|---|
| 1 | 55C | 10 |
| 2 | 56C | 10 |
| 3 | 57C | 10 |
~1 hour and 30 minutes for PCR program

GeneRuler 1kb plus DNA LAdder on 2% gel: 100V, 400amp, 1 hour

GeneRuler 100 bp DNA Ladder on 2% gel: 75V, 400amp, 1 hour
Ladder, 10 original not diluted, 3 bright band diluted to see if there are 2 bands or just one strong band

https://www.thermofisher.com/us/en/home/brands/thermo-scientific/molecular-biology/molecular-biology-learning-center/molecular-biology-resource-library/spotlight-articles/8-DNA-ladder-tips.html
Next steps: PCR at 57C (and maybe 58?) with same samples - original DNA then dilute to 4 ng/uL then PCR and gel again. No dye in ladder and 2% gel like last time.
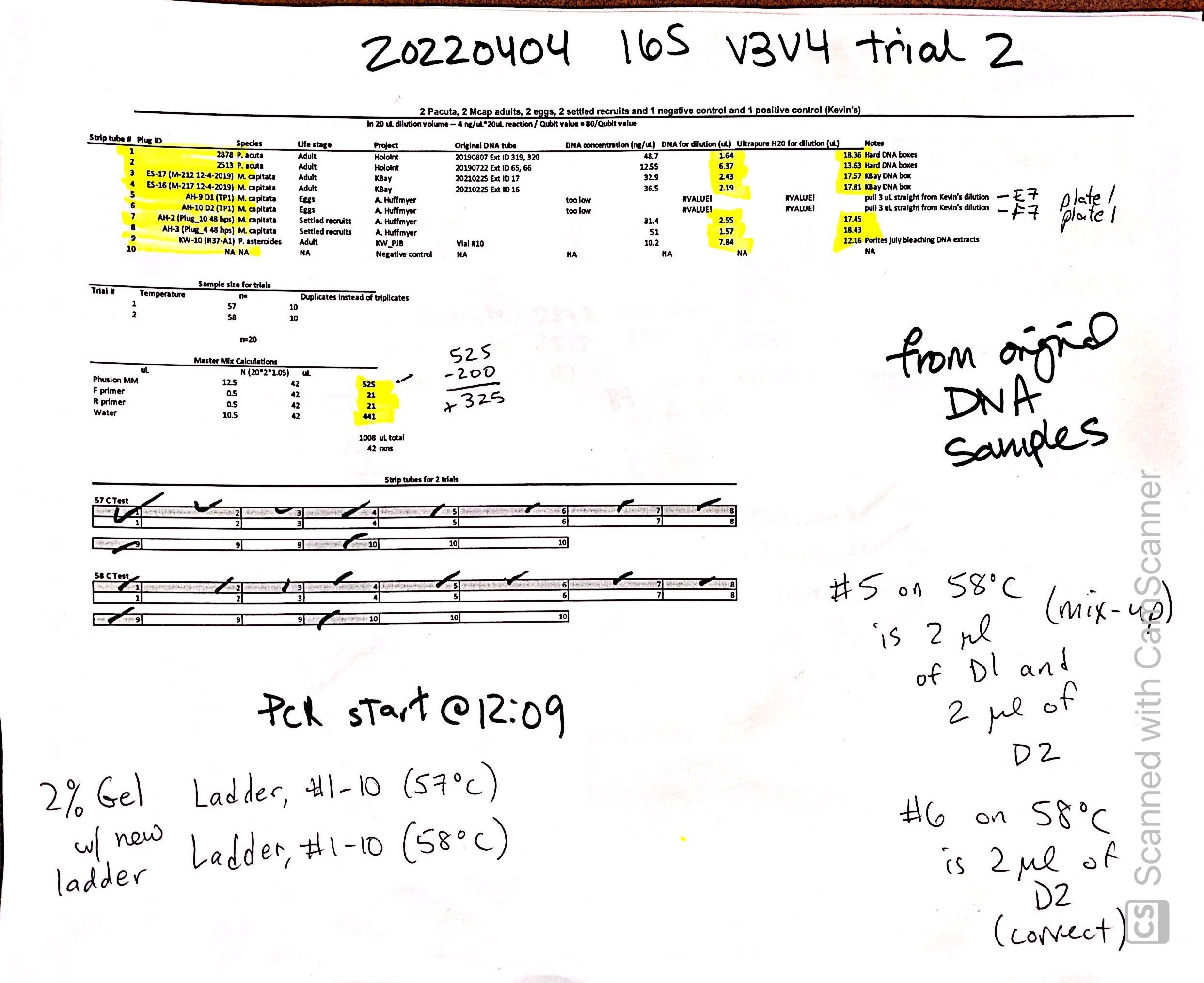
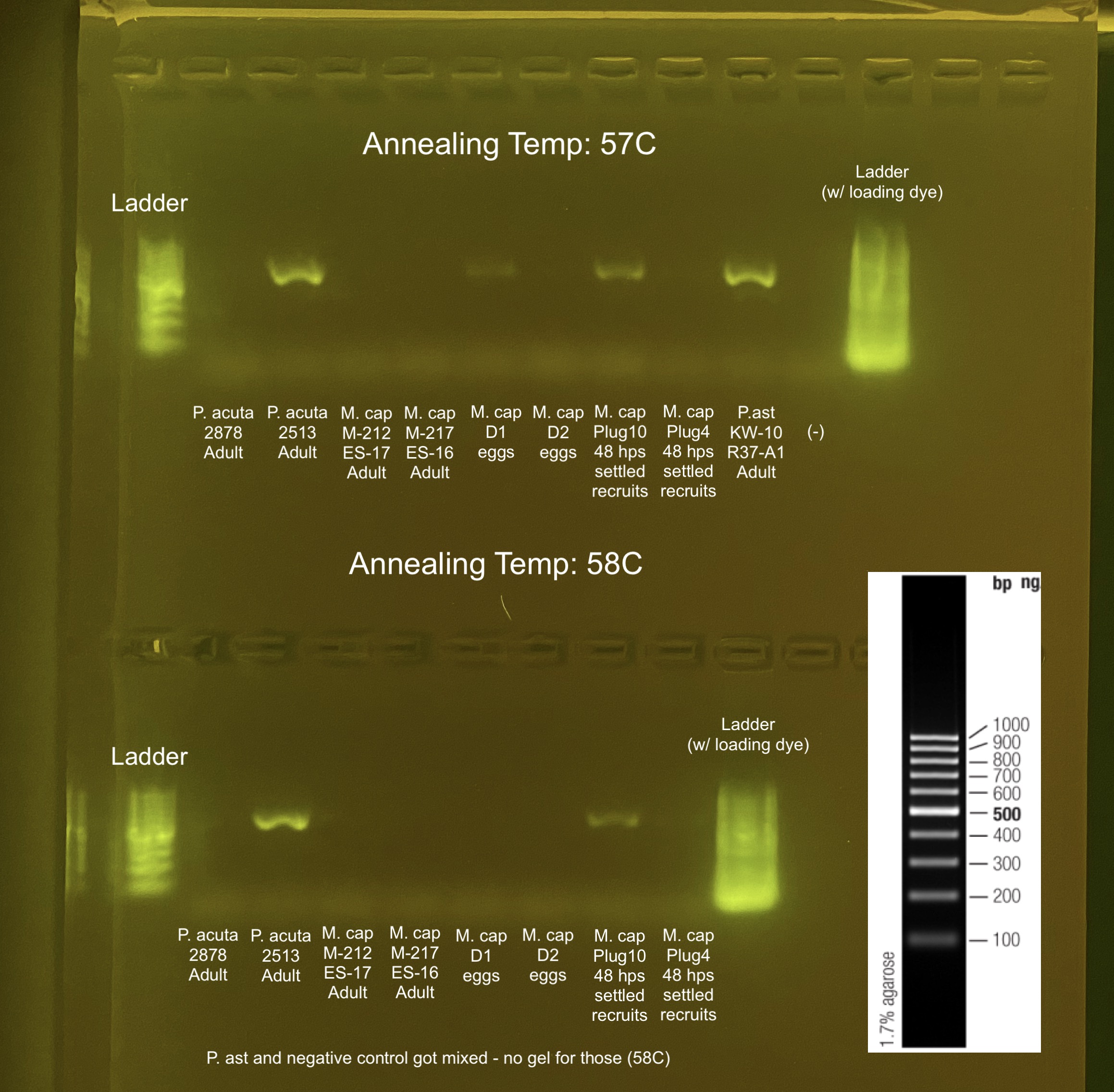
20220404 Trial 2
Same samples as above but re-diluted from original DNA and did duplicates instead of triplicates to save materials. Ran at 57C and 58C to make sure we have the correct annealing temperature. Dilution: in 20 uL dilution volume – 4 ng/uL*20uL reaction / Qubit value = 80/Qubit value.
| Strip tube # | Plug ID | Species | Life stage | Project | Original DNA tube | DNA concentration (ng/uL) | DNA for dilution (uL) | Ultrapure H20 for dilution (uL) | Notes |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2878 | P. acuta | Adult | HoloInt | 20190807 Ext ID 319, 320 | 48.7 | 1.64 | 18.36 | Hard DNA boxes |
| 2 | 2513 | P. acuta | Adult | HoloInt | 20190722 Ext ID 65, 66 | 12.55 | 6.37 | 13.63 | Hard DNA boxes |
| 3 | ES-17 (M-212 12-4-2019) | M. capitata | Adult | KBay | 20210225 Ext ID 17 | 32.9 | 2.43 | 17.57 | KBay DNA box |
| 4 | ES-16 (M-217 12-4-2019) | M. capitata | Adult | KBay | 20210225 Ext ID 16 | 36.5 | 2.19 | 17.81 | KBay DNA box |
| 5 | AH-9 D1 (TP1) | M. capitata | Eggs | A. Huffmyer | too low | #VALUE! | #VALUE! | pull 3 uL straight from Kevin’s dilution | |
| 6 | AH-10 D2 (TP1) | M. capitata | Eggs | A. Huffmyer | too low | #VALUE! | #VALUE! | pull 3 uL straight from Kevin’s dilution | |
| 7 | AH-2 (Plug_10 48 hps) | M. capitata | Settled recruits | A. Huffmyer | 31.4 | 2.55 | 17.45 | ||
| 8 | AH-3 (Plug_4 48 hps) | M. capitata | Settled recruits | A. Huffmyer | 51 | 1.57 | 18.43 | ||
| 9 | KW-10 (R37-A1) | P. asteroides | Adult | KW_PJB | Vial #10 | 10.2 | 7.84 | 12.16 | Porites july bleaching DNA extracts |
| 10 | NA | NA | NA | Negative control | NA | NA | NA | NA | NA |
| Master Mix Calculations | |||
|---|---|---|---|
| uL | N (2021.05) | uL | |
| Phusion MM | 12.5 | 42 | 525 |
| F primer | 0.5 | 42 | 21 |
| R primer | 0.5 | 42 | 21 |
| Water | 10.5 | 42 | 441 |
GeneRuler 100 bp DNA Ladder on 2% gel: 75V, 400amp, 1 hour
20220406 Trial 3
Trial1 = 12 ng DNA input total in that gel band (done in triplicates)
Trial2 = 8 ng DNA input total in that gel band (done in duplicates)
Trial3 - varying DNA input to see if we can get a band to appear in those that don’t have one
Done in triplicates
4 ng/uL already made from Trial 2 - use those
in 20 uL dilution volume – 8 ng/uL*20uL reaction / Qubit value = 160/Qubit value
| SampleID | Input concentration | Original DNA tube | Notes | DNA concentration (ng/uL) | DNA for dilution (uL) | Ultrapure H20 for dilution (uL) | |
|---|---|---|---|---|---|---|---|
| 1 | P.acuta 2513 adult | 4 ng/uL | 20190722 Ext ID 65, 66 | Bright band | 12.55 | 6.37 | 13.63 |
| 2 | P. ast KW-10 | 4 ng/uL | Vial #10 | Bright band | 10.2 | 7.84 | 12.16 |
| 3 | M. cap Plug10 | 4 ng/uL | Labeled plug 10 | Faint/No band | 31.4 | 2.55 | 17.45 |
| 4 | P. acuta 2878 adult | 4 ng/uL | 20190807 Ext ID 319, 320 | Faint/No band | 48.7 | 1.64 | 18.36 |
| 5 | M. cap M-217 | 4 ng/uL | 20210225 Ext ID 16 | No band | 36.5 | 2.19 | 17.81 |
| 6 | M. cap M-212 | 4 ng/uL | 20210225 Ext ID 17 | No band | 32.9 | 2.43 | 17.57 |
| 7 | P.acuta 2513 adult | 8 ng/uL | 20190722 Ext ID 65, 66 | Bright band | 12.55 | 12.75 | 7.25 |
| 8 | P. ast KW-10 | 8 ng/uL | Vial #10 | Bright band | 10.2 | 15.69 | 4.31 |
| 9 | M. cap Plug10 | 8 ng/uL | Labeled plug 10 | Faint/No band | 31.4 | 5.10 | 14.90 |
| 10 | P. acuta 2878 adult | 8 ng/uL | 20190807 Ext ID 319, 320 | Faint/No band | 48.7 | 3.29 | 16.71 |
| 11 | M. cap M-217 | 8 ng/uL | 20210225 Ext ID 16 | No band | 36.5 | 4.38 | 15.62 |
| 12 | M. cap M-212 | 8 ng/uL | 20210225 Ext ID 17 | No band | 32.9 | 4.86 | 15.14 |
| Master Mix Calculations | |||
|---|---|---|---|
| uL | N (1231.05) | uL | |
| Phusion MM | 12.5 | 37.8 | 472.5 |
| F primer | 0.5 | 37.8 | 18.9 |
| R primer | 0.5 | 37.8 | 18.9 |
| Water | 10.5 | 37.8 | 396.9 |

GeneRuler 1kb plus DNA Ladder on 2% gel: 75V, 400amp, 1 hour

Notes
Based on other papers that use 338F and 806R, we have a lot of room to bump up the ng concentrations. Other papers are in the range of 50-100 ng input per sample.
338F and 806R:
Pocillopora - Qin et al 2022
Favites and Acropora - Meenatchi et al 2020
Pocillopora - Yang et al 2020
341F and 802R: Phanerochaete and Galaxea - Cai et al 2018
341F and 785R:
Porites spp. - O’Brien et al 2018
341F and 806R: Nelson’s lab at Hawaii for M. capitata
Adult M. capitata might not work with the 338F primer?
Testing with 341F
20220413 Trial 4
8 ng/uL concentrations except for the two egg samples. 9 coral samples plus 1 negative control to test the new primer 341F.
Resuspend 341F primer concentrate in 584 uL to get 100 uM primer stock.
To get 10 uM primer: 20 uL of the 100 uM stock + 180 uL of Ultrapure water.
| PCR strip tube # | SampleID | Input concentration | DNA location | Result from 338F trials | Qubit concentration | DNA (uL) | H20 (uL) |
|---|---|---|---|---|---|---|---|
| 1 | P.acuta 2513 adult | 8 ng/uL | 20220406 8 ng/uL dilution strip tube (tube 7) | Bright band | |||
| 2 | P. acuta 2878 adult | 8 ng/uL | 20220406 8 ng/uL dilution strip tube (tube 10) | Faint/No band | |||
| 3 | M. cap M-217 | 8 ng/uL | 20220406 8 ng/uL dilution strip tube (tube 11) | No band | |||
| 4 | M. cap M-212 | 8 ng/uL | 20220406 8 ng/uL dilution strip tube (tube 12) | No band | |||
| 5 | M. cap Plug10 | 8 ng/uL | 20220406 8 ng/uL dilution strip tube (tube 9) | Faint/No band | |||
| 6 | M. cap Plug4 | 8 ng/uL | Labeled Plug4 in AH’s freezer box | No band | 51 | 3.14 | 16.86 |
| 7 | D1 eggs | too low | KW 4ng/uL plate 1 well E7 | Faint/No band | |||
| 8 | D2 eggs | too low | KW 4ng/uL plate 1 well F7 | Faint/No band | |||
| 9 | P. ast KW-10 | 8 ng/uL | 20220406 8 ng/uL dilution strip tube | Bright band | |||
| 10 | Negative control | NA | NA | NA | NA | NA | NA |
| Master Mix Calculations | |||
|---|---|---|---|
| uL | N (1031.05) | uL | |
| Phusion MM | 12.5 | 31.5 | 393.75 |
| F primer | 0.5 | 31.5 | 15.75 |
| R primer | 0.5 | 31.5 | 15.75 |
| Water | 10.5 | 31.5 | 330.75 |

GeneRuler 1kb plus DNA Ladder on 2% gel: 75V, 400amp, 1 hour

Higher Annealing Temperature Test
20220418 Trial 5
DNA BR Standard 1: 198.05
DNA BR Standard 2: 21,549.69
M-217: 14.8 14.3
M-212: 29.0; 28.6
There is DNA for both M.cap adults that don’t work with the V3 primer…
Testing higher annealing temperature using 338F - Trial 5
4 ng/uL to be able to compare to 56, 57, and 58 C trials
| PCR strip tube # | SampleID | Input concentration | DNA location | Annealing Temperature |
|---|---|---|---|---|
| 1 | P. acuta 2513 | 4 ng/uL | 20220404 strip tube 4 ng/uL | 60 C |
| 2 | P. ast KW-10 | 4 ng/uL | 20220404 strip tube 4 ng/uL | 60 C |
| 3 | Plug 10 settled recruit | 4 ng/uL | 20220404 strip tube 4 ng/uL | 60 C |
| 4 | D1 eggs | too follow | Original DNA tube - 3 uL | 60 C |
| 5 | M-217 | 4 ng/uL | 20220404 strip tube 4 ng/uL | 60 C |
| 6 | negative control | NA | NA | 60 C |
| 7 | P. acuta 2513 | 4 ng/uL | 20220404 strip tube 4 ng/uL | 63 C |
| 8 | P. ast KW-10 | 4 ng/uL | 20220404 strip tube 4 ng/uL | 63 C |
| 9 | Plug 10 settled recruit | 4 ng/uL | 20220404 strip tube 4 ng/uL | 63 C |
| 10 | D1 eggs | too follow | Original DNA tube - 3 uL | 63 C |
| 11 | M-217 | 4 ng/uL | 20220404 strip tube 4 ng/uL | 63 C |
| 12 | negative control | NA | NA | 63 C |
| uL | N (1231.05) | uL | |
|---|---|---|---|
| Phusion MM | 12.5 | 37.8 | 472.5 |
| F primer | 0.5 | 37.8 | 18.9 |
| R primer | 0.5 | 37.8 | 18.9 |
| Water | 10.5 | 37.8 | 396.9 |

GeneRuler 1kb plus DNA Ladder on 2% gel: 75V, 400amp, 1 hour

Annealing Temperature comparison
all 4 ng/uL input with 338F and 806RB primers. All temperature seem to work..

20220419 Trial 6
Testing a couple M. capitata samples with 515F again to figure out if it’s a V3 region issue or DNA sample issue.
8 ng/uL DNA input for 2 temperatures
| PCR strip tube # | SampleID | Input concentration | DNA location | Annealing Temperature |
|---|---|---|---|---|
| 1 | M-217 (Ext ID 16) | 8 ng/uL | 20220406 8 ng/uL dilution strip tube | 57 C |
| 2 | M-212 (Ext ID 17) | 8 ng/uL | 20220406 8 ng/uL dilution strip tube | 57 C |
| 3 | P. ast KW-10 | 8 ng/uL | 20220406 8 ng/uL dilution strip tube | 57 C |
| 4 | Negative control | 8 ng/uL | Original DNA tube - 3 uL | 57 C |
| 5 | M-217 (Ext ID 16) | 8 ng/uL | 20220406 8 ng/uL dilution strip tube | 63 C |
| 6 | M-212 (Ext ID 17) | 8 ng/uL | 20220406 8 ng/uL dilution strip tube | 63 C |
| 7 | P. ast KW-10 | 8 ng/uL | 20220406 8 ng/uL dilution strip tube | 63 C |
| 8 | Negative control | 8 ng/uL | Original DNA tube - 3 uL | 63 C |
| uL | N (831.05) | uL | |
|---|---|---|---|
| Phusion MM | 12.5 | 25.2 | 315 |
| 515 F primer | 0.5 | 25.2 | 12.6 |
| 806 RB primer | 0.5 | 25.2 | 12.6 |
| Water | 10.5 | 25.2 | 264.6 |

GeneRuler 1kb plus DNA Ladder on 2% gel: 75V, 400amp, 1 hour

M. cap adults are not working with V3 region specifically, both 341F and 338F primers.
Sequencing test run 1
Questions to answer from sequencing:
- Do we get microbiome or host sequences from 338F and 341F V3V4 region PCR products?
- Which V3V34 primer (338F and or 341F) yields higher microbiome sequences?
- Does 63C annealing temperature for the V4 (515F) region yield better microbiome vs host ratios? Is this still all host?
- What kind of noise do we get from negative controls?
19 samples to sequence
| SampleID | Strip tube # to Janet | 16S Region | Forward Primer Used | Annealing Temperature | SampleID | DNA input | Date PCR run | Band information | Replicate # (uL total after gel) | uL leftover after |
|---|---|---|---|---|---|---|---|---|---|---|
| ELS001 | 1 | V3V4 | 338F | 57 C | P. acuta 2513 adult | 8 ng/uL (24 ng total) | 20220406 Trial 3 | Bright | 3 | 45 |
| ELS002 | 2 | V3V4 | 338F | 57 C | P. ast KW-10 adult | 8 ng/uL (24 ng total) | 20220406 Trial 3 | Bright | 3 | 45 |
| ELS003 | 3 | V3V4 | 338F | 57 C | Plug 10 settled recruit | 8 ng/uL (24 ng total) | 20220406 Trial 3 | Bright | 3 | 45 |
| ELS004 | 4 | V3V4 | 338F | 57 C | P. acuta 2878 | 4 ng/uL (12 ng total) | 20220321 Trial 2 | Faint | 2 | 20 |
| ELS005 | 5 | V3V4 | 338F | 57 C | D1 eggs | too low | 20220321 Trial 2 | Faint | 2 | 20 |
| ELS006 | 6 | V3V4 | 338F | 63 C | P. acuta 2513 adult | 4 ng/uL (12 ng total) | 20220418 Trial 5 | Bright | 3 | 45 |
| ELS007 | 7 | V3V4 | 338F | 63 C | P. ast KW-10 adult | 4 ng/uL (12 ng total) | 20220418 Trial 5 | Bright | 3 | 45 |
| ELS008 | 8 | V3V4 | 338F | 63 C | Plug 10 settled recruit | 4 ng/uL (12 ng total) | 20220418 Trial 5 | Faint | 3 | 45 |
| ELS009 | 9 | V3V4 | 341F | 57 C | P. acuta 2513 adult | 8 ng/uL (24 ng total) | 20220413 Trial 4 | Bright | 3 | 45 |
| ELS010 | 10 | V3V4 | 341F | 57 C | P. ast KW-10 adult | 8 ng/uL (24 ng total) | 20220413 Trial 4 | Bright | 3 | 45 |
| ELS011 | 11 | V3V4 | 341F | 57 C | Plug 10 settled recruit | 8 ng/uL (24 ng total) | 20220413 Trial 4 | Faint | 3 | 45 |
| ELS012 | 12 | V3V4 | 341F | 57 C | Plug 4 settled recruit | 8 ng/uL (24 ng total) | 20220413 Trial 4 | Faint | 3 | 45 |
| ELS013 | 13 | V3V4 | 341F | 57 C | D1 eggs | too low | 20220413 Trial 4 | Very faint | 3 | 45 |
| ELS014 | 14 | V4 | 515F | 63 C | M-217 | 8 ng/uL (24 ng total) | 20220419 Trial 6 | Bright | 3 | 45 |
| ELS015 | 15 | V4 | 515F | 63 C | M-212 | 8 ng/uL (24 ng total) | 20220419 Trial 6 | Bright | 3 | 45 |
| ELS016 | 16 | V4 | 515F | 63 C | P. ast KW-10 adult | 8 ng/uL (24 ng total) | 20220419 Trial 6 | Bright | 3 | 45 |
| ELS017 | 17 | V3V4 | 338F | 57 C | Negative control | NA | 20220406 Trial 3 | None | 3 | 45 |
| ELS018 | 18 | V3V4 | 341F | 57 C | Negative control | NA | 20220413 Trial 4 | None | 3 | 45 |
| ELS019 | 19 | V4 | 515F | 63 C | Negative control | NA | 20220419 Trial 6 | None | 3 | 45 |
Stitched gel image from the above samples


